Related Research Articles
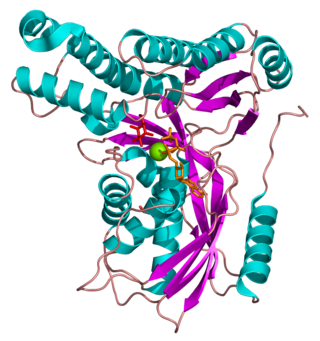
Galactokinase is an enzyme (phosphotransferase) that facilitates the phosphorylation of α-D-galactose to galactose 1-phosphate at the expense of one molecule of ATP. Galactokinase catalyzes the second step of the Leloir pathway, a metabolic pathway found in most organisms for the catabolism of α-D-galactose to glucose 1-phosphate. First isolated from mammalian liver, galactokinase has been studied extensively in yeast, archaea, plants, and humans.
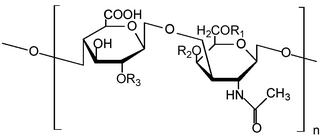
Glycosaminoglycans (GAGs) or mucopolysaccharides are long, linear polysaccharides consisting of repeating disaccharide units. The repeating two-sugar unit consists of a uronic sugar and an amino sugar, except in the case of the sulfated glycosaminoglycan keratan, where, in place of the uronic sugar there is a galactose unit. GAGs are found in vertebrates, invertebrates and bacteria. Because GAGs are highly polar molecules and attract water; the body uses them as lubricants or shock absorbers.

Heparan sulfate (HS) is a linear polysaccharide found in all animal tissues. It occurs as a proteoglycan in which two or three HS chains are attached in close proximity to cell surface or extracellular matrix proteins. In this form, HS binds to a variety of protein ligands, including Wnt, and regulates a wide range of biological activities, including developmental processes, angiogenesis, blood coagulation, abolishing detachment activity by GrB, and tumour metastasis. HS has also been shown to serve as cellular receptor for a number of viruses, including the respiratory syncytial virus. One study suggests that cellular heparan sulfate has a role in SARS-CoV-2 Infection, particularly when the virus attaches with ACE2.

Galactosyltransferase is a type of glycosyltransferase which catalyzes the transfer of galactose. An example is B-N-acetylglucosaminyl-glycopeptide b-1,4-galactosyltransferase.

A disintegrin and metalloproteinase with thrombospondin motifs 4 is an enzyme that in humans is encoded by the ADAMTS4 gene.
In enzymology, a xylosylprotein 4-beta-galactosyltransferase is an enzyme that catalyzes the chemical reaction

Beta-1,4-galactosyltransferase 7 also known as galactosyltransferase I is an enzyme that in humans is encoded by the B4GALT7 gene. Galactosyltransferase I catalyzes the synthesis of the glycosaminoglycan-protein linkage in proteoglycans. Proteoglycans in turn are structural components of the extracellular matrix that is found between cells in connective tissues.

Xylosyltransferase 2 is an enzyme that in humans is encoded by the XYLT2 gene.
Lambda-carrageenase is an enzyme which breaks down a polysaccharide found in red seaweeds, lambda-carrageenan. This enzyme has only been found in marine bacteria.

In molecular biology, a carbohydrate-binding module (CBM) is a protein domain found in carbohydrate-active enzymes. The majority of these domains have carbohydrate-binding activity. Some of these domains are found on cellulosomal scaffoldin proteins. CBMs were previously known as cellulose-binding domains. CBMs are classified into numerous families, based on amino acid sequence similarity. There are currently 64 families of CBM in the CAZy database.
In molecular biology, glycoside hydrolase family 44 is a family of glycoside hydrolases.
N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase is an enzyme with systematic name 3'-phosphoadenylyl-sulfate:(dermatan)-4-O-sulfo-N-acetyl-D-galactosamine 6-O-sulfotransferase. This enzyme catalyses the following chemical reaction
Dermatan 4-sulfotransferase is an enzyme with systematic name 3'-phospho-5'-adenylyl sulfate:(dermatan)-N-acetyl-D-galactosamine 4-sulfotransferase. This enzyme catalyses the following chemical reaction
κ-Carrageenase is an enzyme with systematic name κ-carrageenan 4-β-D-glycanohydrolase (configuration-retaining). It catalyses the endohydrolysis of (1→4)-β-D-linkages between D-galactose 4-sulfate and 3,6-anhydro-D-galactose in κ-carrageenans
Heparanase is an enzyme with systematic name heparan sulfate N-sulfo-D-glucosamine endoglucanase. This enzyme catalyses the following chemical reaction
Beta porphyranase is an enzyme responsible for the degradation of porphyran, which composes the cell wall of red algae. So far only five β-porphyranases have been identified: PorA and PorB are found in the marine bacteria Zobellia galactinovirans. A wild-type porphyranase activity has been found in Pseudoalteromonas atlantica. BpGH16B and BpGH86A have been found in the human gut bacterium, Bacteroides plebeius, of Japanese individuals.
Gellan tetrasaccharide unsaturated glucuronyl hydrolase (EC 3.2.1.179, UGL, unsaturated glucuronyl hydrolase) is an enzyme with systematic name beta-D-4-deoxy-Delta4-GlcAp-(1->4)-beta-D-Glcp-(1->4)-alpha-L-Rhap-(1->3)-beta-D-Glcp beta-D-4-deoxy-Delta4-GlcAp hydrolase. This enzyme catalyses the following chemical reaction

Gideon John Davies is a Professor of Chemistry in the Structural Biology Laboratory (YSBL) at the University of York, UK. Davies is best known for his ground-breaking studies into carbohydrate-active enzymes, notably analysing the conformational and mechanistic basis for catalysis and applying this for societal benefit. In 2016 Davies was apppointed the Royal Society Ken Murray Research Professor at the University of York.
Zobellia galactanivorans is a gram-negative marine bacterium isolated from the surface of red algae of the coast of France. Z. galactanivorans forms yellow colonies with a bacillus or diplobacillus morphology. Furthermore, it is mesophilic and can grow degrade carrageenans and agars - both found in the cell wall of red algae. Z. galactanivorans contains the gene porA and porB, each encoding a β-porphyranase.
Ulvan lyase is an enzyme found within the cell-wall of the marine organism Ulvales, and some marine bacterium. A lyase is a class of enzyme that catalyzes the breakdown of chemical bonds through an elimination reaction mechanism, rather than a substitution reaction mechanism. Ulvan lyase belongs to the polysaccharide lyase family, a type of enzyme that primarily functions to cleave glycosidic linkages in polysaccharides.
References
- ↑ Barbeyron T, Michel G, Potin P, Henrissat B, Kloareg B (November 2000). "iota-Carrageenases constitute a novel family of glycoside hydrolases, unrelated to that of kappa-carrageenases". The Journal of Biological Chemistry. 275 (45): 35499–505. doi: 10.1074/jbc.m003404200 . PMID 10934194.
- ↑ Michel G, Chantalat L, Fanchon E, Henrissat B, Kloareg B, Dideberg O (October 2001). "The iota-carrageenase of Alteromonas fortis. A beta-helix fold-containing enzyme for the degradation of a highly polyanionic polysaccharide". The Journal of Biological Chemistry. 276 (43): 40202–9. doi: 10.1074/jbc.M100670200 . PMID 11493601.
- ↑ Michel G, Helbert W, Kahn R, Dideberg O, Kloareg B (November 2003). "The structural bases of the processive degradation of iota-carrageenan, a main cell wall polysaccharide of red algae". Journal of Molecular Biology. 334 (3): 421–33. doi:10.1016/j.jmb.2003.09.056. PMID 14623184.