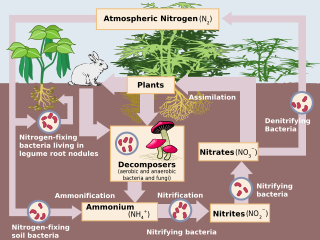
Nitrification is the biological oxidation of ammonia to nitrate via the intermediary nitrite. Nitrification is an important step in the nitrogen cycle in soil. The process of complete nitrification may occur through separate organisms or entirely within one organism, as in comammox bacteria. The transformation of ammonia to nitrite is usually the rate limiting step of nitrification. Nitrification is an aerobic process performed by small groups of autotrophic bacteria and archaea.

Acidobacteriota is a phylum of Gram-negative bacteria. Its members are physiologically diverse and ubiquitous, especially in soils, but are under-represented in culture.

Anammox, an abbreviation for "anaerobic ammonium oxidation", is a globally important microbial process of the nitrogen cycle that takes place in many natural environments. The bacteria mediating this process were identified in 1999, and were a great surprise for the scientific community. In the anammox reaction, nitrite and ammonium ions are converted directly into diatomic nitrogen and water.
Methanotrophs are prokaryotes that metabolize methane as their source of carbon and chemical energy. They are bacteria or archaea, can grow aerobically or anaerobically, and require single-carbon compounds to survive.

Sphingomonadaceae are a gram-negative bacterial family of the Alphaproteobacteria. An important feature is the presence of sphingolipids in the outer membrane of the cell wall. The cells are ovoid or rod-shaped. Others are also pleomorphic, i.e. the cells change the shape over time. Some species from Sphingomonadaceae family are dominant components of biofilms.
Paracoccus denitrificans, is a coccoid bacterium known for its nitrate reducing properties, its ability to replicate under conditions of hypergravity and for being a relative of the eukaryotic mitochondrion.
Nitrospira translate into “a nitrate spiral” is a genus of bacteria within the monophyletic clade of the Nitrospirota phylum. The first member of this genus was described 1986 by Watson et al., isolated from the Gulf of Maine. The bacterium was named Nitrospira marina. Populations were initially thought to be limited to marine ecosystems, but it was later discovered to be well-suited for numerous habitats, including activated sludge of wastewater treatment systems, natural biological marine settings, water circulation biofilters in aquarium tanks, terrestrial systems, fresh and salt water ecosystems, agricultural lands and hot springs. Nitrospira is a ubiquitous bacterium that plays a role in the nitrogen cycle by performing nitrite oxidation in the second step of nitrification. Nitrospira live in a wide array of environments including but not limited to, drinking water systems, waste treatment plants, rice paddies, forest soils, geothermal springs, and sponge tissue. Despite being abundant in many natural and engineered ecosystems Nitrospira are difficult to culture, so most knowledge of them is from molecular and genomic data. However, due to their difficulty to be cultivated in laboratory settings, the entire genome was only sequenced in one species, Nitrospira defluvii. In addition, Nitrospira bacteria's 16S rRNA sequences are too dissimilar to use for PCR primers, thus some members go unnoticed. In addition, members of Nitrospira with the capabilities to perform complete nitrification has also been discovered and cultivated.
Gammaproteobacteria is a class of bacteria in the phylum Pseudomonadota. It contains about 250 genera, which makes it the most genus-rich taxon of the Prokaryotes. Several medically, ecologically, and scientifically important groups of bacteria belong to this class. All members of this class are Gram-negative. It is the most phylogenetically and physiologically diverse class of the Pseudomonadota.
In taxonomy, Stappia is a genus of the Hyphomicrobiales. Some members of the genus oxidize carbon monoxide (CO) aerobically. Stappia indica is a diatom associated bacterium which is known to inhibit the growth of diatoms such as Thalassiosira pseudonana.

The Nitrososphaerota are a phylum of the Archaea proposed in 2008 after the genome of Cenarchaeum symbiosum was sequenced and found to differ significantly from other members of the hyperthermophilic phylum Thermoproteota. Three described species in addition to C. symbiosum are Nitrosopumilus maritimus, Nitrososphaera viennensis, and Nitrososphaera gargensis. The phylum was proposed in 2008 based on phylogenetic data, such as the sequences of these organisms' ribosomal RNA genes, and the presence of a form of type I topoisomerase that was previously thought to be unique to the eukaryotes. This assignment was confirmed by further analysis published in 2010 that examined the genomes of the ammonia-oxidizing archaea Nitrosopumilus maritimus and Nitrososphaera gargensis, concluding that these species form a distinct lineage that includes Cenarchaeum symbiosum. The lipid crenarchaeol has been found only in Nitrososphaerota, making it a potential biomarker for the phylum. Most organisms of this lineage thus far identified are chemolithoautotrophic ammonia-oxidizers and may play important roles in biogeochemical cycles, such as the nitrogen cycle and the carbon cycle. Metagenomic sequencing indicates that they constitute ~1% of the sea surface metagenome across many sites.
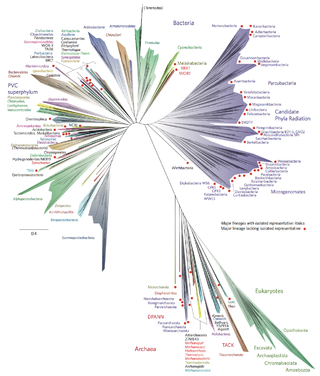
Bacterial phyla constitute the major lineages of the domain Bacteria. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.
Saccharibacteria, formerly known as TM7, is a major bacterial lineage. It was discovered through 16S rRNA sequencing.

2,4-Diacetylphloroglucinol or Phl is a natural phenol found in several bacteria:

The class Zetaproteobacteria is the sixth and most recently described class of the Pseudomonadota. Zetaproteobacteria can also refer to the group of organisms assigned to this class. The Zetaproteobacteria were originally represented by a single described species, Mariprofundus ferrooxydans, which is an iron-oxidizing neutrophilic chemolithoautotroph originally isolated from Kamaʻehuakanaloa Seamount in 1996 (post-eruption). Molecular cloning techniques focusing on the small subunit ribosomal RNA gene have also been used to identify a more diverse majority of the Zetaproteobacteria that have as yet been unculturable.
Community fingerprinting is a set of molecular biology techniques that can be used to quickly profile the diversity of a microbial community. Rather than directly identifying or counting individual cells in an environmental sample, these techniques show how many variants of a gene are present. In general, it is assumed that each different gene variant represents a different type of microbe. Community fingerprinting is used by microbiologists studying a variety of microbial systems to measure biodiversity or track changes in community structure over time. The method analyzes environmental samples by assaying genomic DNA. This approach offers an alternative to microbial culturing, which is important because most microbes cannot be cultured in the laboratory. Community fingerprinting does not result in identification of individual microbe species; instead, it presents an overall picture of a microbial community. These methods are now largely being replaced by high throughput sequencing, such as targeted microbiome analysis and metagenomics.
Nitrososphaera is a mesophilic genus of ammonia-oxidizing Crenarchaeota. The first Nitrososphaera organism was discovered in garden soils at the University of Vienna leading to the categorization of a new genus, family, order and class of Archaea. This genus is contains three distinct species: N. viennensis, Ca. N. gargensis, and Ca N. evergladensis. Nitrososphaera are chemolithoautotrophs and have important biogeochemical roles as nitrifying organisms.

The root microbiome is the dynamic community of microorganisms associated with plant roots. Because they are rich in a variety of carbon compounds, plant roots provide unique environments for a diverse assemblage of soil microorganisms, including bacteria, fungi, and archaea. The microbial communities inside the root and in the rhizosphere are distinct from each other, and from the microbial communities of bulk soil, although there is some overlap in species composition.

Bioluminescent bacteria are light-producing bacteria that are predominantly present in sea water, marine sediments, the surface of decomposing fish and in the gut of marine animals. While not as common, bacterial bioluminescence is also found in terrestrial and freshwater bacteria. These bacteria may be free living or in symbiosis with animals such as the Hawaiian Bobtail squid or terrestrial nematodes. The host organisms provide these bacteria a safe home and sufficient nutrition. In exchange, the hosts use the light produced by the bacteria for camouflage, prey and/or mate attraction. Bioluminescent bacteria have evolved symbiotic relationships with other organisms in which both participants benefit close to equally. Another possible reason bacteria use luminescence reaction is for quorum sensing, an ability to regulate gene expression in response to bacterial cell density.
Gabriele Berg is a biologist, biotechnologist and university lecturer in Environmental and Ecological Technology at the Technical University of Graz. Her research emphasis is on the development of sustainable methods of plant vitalisation with Bioeffectors and molecular analysis of microbial processes in the soil, particularly in the Rhizosphere.
Hydrocarbonoclastic bacteria are a heterogeneous group of prokaryotes which can degrade and utilize hydrocarbon compounds as source of carbon and energy. Despite being present in most of environments around the world, several of these specialized bacteria live in the sea and have been isolated from polluted seawater.












