Related Research Articles

A genetically modified organism (GMO) is any organism whose genetic material has been altered using genetic engineering techniques. The exact definition of a genetically modified organism and what constitutes genetic engineering varies, with the most common being an organism altered in a way that "does not occur naturally by mating and/or natural recombination". A wide variety of organisms have been genetically modified (GM), from animals to plants and microorganisms. Genes have been transferred within the same species, across species, and even across kingdoms. New genes can be introduced, or endogenous genes can be enhanced, altered, or knocked out.

Genetic engineering, also called genetic modification or genetic manipulation, is the modification and manipulation of an organism's genes using technology. It is a set of technologies used to change the genetic makeup of cells, including the transfer of genes within and across species boundaries to produce improved or novel organisms. New DNA is obtained by either isolating and copying the genetic material of interest using recombinant DNA methods or by artificially synthesising the DNA. A construct is usually created and used to insert this DNA into the host organism. The first recombinant DNA molecule was made by Paul Berg in 1972 by combining DNA from the monkey virus SV40 with the lambda virus. As well as inserting genes, the process can be used to remove, or "knock out", genes. The new DNA can be inserted randomly, or targeted to a specific part of the genome.

Take-all is a plant disease affecting the roots of grass and cereal plants in temperate climates caused by the fungus Gaeumannomyces tritici. All varieties of wheat and barley are susceptible. It is an important disease in winter wheat in Western Europe particularly, and is favoured by conditions of intensive production and monoculture.

https://canto.phi-base.org/PHI-baseFile:PHI-base+01.jpgContentDescriptionPathogen-Host+Interactions+databaseData+typescapturedphenotypes+of+microbial+mutantsOrganisms~280+fungal,+bacterial+and+protist+pathogens+of+agronomic+and+medical+importance+tested+on+~230+hostsContactResearch+centerRothamsted+ResearchPrimary+citationPMID 31733065Release+dateMay+2005AccessData+formatXML,+FASTAWebsitePHI-baseToolsWebPHI-base+SearchPHIB-BLASTPHI-Canto+(Author+curation)MiscellaneousLicenseCreative+Commons+Attribution-NoDerivatives+4.0+International+LicenseVersioningYesData+releasefrequency6+monthlyVersion4.15+(May+2023)Curation+policyManual+Curation

Hypersensitive response (HR) is a mechanism used by plants to prevent the spread of infection by microbial pathogens. HR is characterized by the rapid death of cells in the local region surrounding an infection and it serves to restrict the growth and spread of pathogens to other parts of the plant. It is analogous to the innate immune system found in animals, and commonly precedes a slower systemic response, which ultimately leads to systemic acquired resistance (SAR). HR can be observed in the vast majority of plant species and is induced by a wide range of plant pathogens such as oomycetes, viruses, fungi and even insects.
The gene-for-gene relationship was discovered by Harold Henry Flor who was working with rust (Melampsora lini) of flax (Linum usitatissimum). Flor showed that the inheritance of both resistance in the host and parasite ability to cause disease is controlled by pairs of matching genes. One is a plant gene called the resistance (R) gene. The other is a parasite gene called the avirulence (Avr) gene. Plants producing a specific R gene product are resistant towards a pathogen that produces the corresponding Avr gene product. Gene-for-gene relationships are a widespread and very important aspect of plant disease resistance. Another example can be seen with Lactuca serriola versus Bremia lactucae.

Genetically modified plants have been engineered for scientific research, to create new colours in plants, deliver vaccines, and to create enhanced crops. Plant genomes can be engineered by physical methods or by use of Agrobacterium for the delivery of sequences hosted in T-DNA binary vectors. Many plant cells are pluripotent, meaning that a single cell from a mature plant can be harvested and then under the right conditions form a new plant. This ability is most often taken advantage by genetic engineers through selecting cells that can successfully be transformed into an adult plant which can then be grown into multiple new plants containing transgene in every cell through a process known as tissue culture.
Jeffery Lee Dangl is an American biologist. He is currently John N. Couch Professor of Biology at the University of North Carolina at Chapel Hill.

Serine/threonine-protein kinase PAK 1 is an enzyme that in humans is encoded by the PAK1 gene.

RE1-Silencing Transcription factor (REST), also known as Neuron-Restrictive Silencer Factor (NRSF), is a protein which in humans is encoded by the REST gene, and acts as a transcriptional repressor. REST is expressly involved in the repression of neural genes in non-neuronal cells. Many genetic disorders have been tied to alterations in the REST expression pattern, including colon and small-cell lung carcinomas found with truncated versions of REST. In addition to these cancers, defects in REST have also been attributed a role in Huntington Disease, neuroblastomas, and the effects of epileptic seizures and ischemia.

Plant disease resistance protects plants from pathogens in two ways: by pre-formed structures and chemicals, and by infection-induced responses of the immune system. Relative to a susceptible plant, disease resistance is the reduction of pathogen growth on or in the plant, while the term disease tolerance describes plants that exhibit little disease damage despite substantial pathogen levels. Disease outcome is determined by the three-way interaction of the pathogen, the plant and the environmental conditions.

In the fields of molecular biology and genetics, a pan-genome is the entire set of genes from all strains within a clade. More generally, it is the union of all the genomes of a clade. The pan-genome can be broken down into a "core pangenome" that contains genes present in all individuals, a "shell pangenome" that contains genes present in two or more strains, and a "cloud pangenome" that contains genes only found in a single strain. Some authors also refer to the cloud genome as "accessory genome" containing 'dispensable' genes present in a subset of the strains and strain-specific genes. Note that the use of the term 'dispensable' has been questioned, at least in plant genomes, as accessory genes play "an important role in genome evolution and in the complex interplay between the genome and the environment". The field of study of the pangenome is called pangenomics.

Detlef Weigel is a German American scientist working at the interface of developmental and evolutionary biology.

A genetically modified tomato, or transgenic tomato, is a tomato that has had its genes modified, using genetic engineering. The first trial genetically modified food was a tomato engineered to have a longer shelf life, which was on the market briefly beginning on May 21, 1994. The first direct consumption tomato was approved in Japan in 2021. Primary work is focused on developing tomatoes with new traits like increased resistance to pests or environmental stresses. Other projects aim to enrich tomatoes with substances that may offer health benefits or be more nutritious. As well as aiming to produce novel crops, scientists produce genetically modified tomatoes to understand the function of genes naturally present in tomatoes.
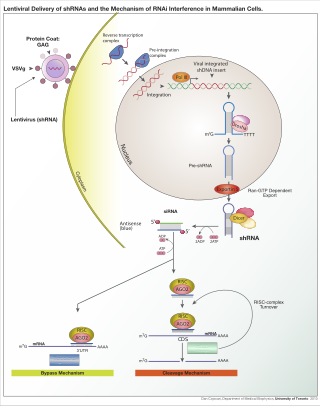
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents.
Nicholas Paul Harberd is Sibthorpian Professor of Plant Science and former head of the Department of Plant Sciences at the University of Oxford, and Fellow of St John's College, Oxford.
In plant biology, elicitors are extrinsic or foreign molecules often associated with plant pests, diseases or synergistic organisms. Elicitor molecules can attach to special receptor proteins located on plant cell membranes. These receptors are able to recognize the molecular pattern of elicitors and trigger intracellular defence signalling via the octadecanoid pathway. This response results in the enhanced synthesis of metabolites which reduce damage and increase resistance to pest, disease or environmental stress. This is an immune response called pattern triggered immunity (PTI).

Catherine Rosemary Martin is a Professor of Plant Sciences at the University of East Anglia (UEA) and project leader at the John Innes Centre, Norwich, co-ordinating research into the relationship between diet and health and how crops can be fortified to improve diets and address escalating chronic disease globally.

Liam Dolan is a Senior Group Leader at the Gregor Mendel Institute of Molecular Plant Biology (GMI) of the Austrian Academy of Sciences, the Sherardian Professor of Botany in the Department of Biology at the University of Oxford and a Fellow of Magdalen College, Oxford.

Nicholas José Talbot FRS FRSB is Group Leader and Executive Director at The Sainsbury Laboratory in Norwich.
References
- 1 2 3 4 5 6 7 Anon (1998). "Jones, Jonathan Dallas George" . Who's Who (online Oxford University Press ed.). Oxford: A & C Black. doi:10.1093/ww/9780199540884.013.U22396.(Subscription or UK public library membership required.)
- 1 2 3 Anon (2003). "Professor Jonathan Jones FRS". royalsociety.org. London: Royal Society. One or more of the preceding sentences incorporates text from the royalsociety.org website where:
“All text published under the heading 'Biography' on Fellow profile pages is available under Creative Commons Attribution 4.0 International License.” --Royal Society Terms, conditions and policies at the Wayback Machine (archived 2016-11-11)
- 1 2 Viegas, Jennifer (2018). "Profile of Jonathan D. G. Jones". Proceedings of the National Academy of Sciences. 115 (41): 10191–10194. Bibcode:2018PNAS..11510191V. doi: 10.1073/pnas.1815072115 . ISSN 0027-8424. PMC 6187187 . PMID 30249645.
- 1 2 "Jonathan Jones". www.nasonline.org.
- 1 2 Jonathan D. G. Jones publications indexed by Google Scholar
- 1 2 Bedbrook, J. R.; Jones, J.; O'Dell, M.; Thompson, R. D.; Flavell, R. B. (1980). "A molecular description of telometic heterochromatin in secale species". Cell. 19 (2): 545–560. doi:10.1016/0092-8674(80)90529-2. PMID 6244112. S2CID 31566816.
- ↑ Jonathan D. G. Jones's publications indexed by the Scopus bibliographic database. (subscription required)
- ↑ Hammond-Kosack, K. E.; Silverman, P.; Raskin, I.; Jones, J. (1996). "Race-Specific Elicitors of Cladosporium fulvum Induce Changes in Cell Morphology and the Synthesis of Ethylene and Salicylic Acid in Tomato Plants Carrying the Corresponding Cf Disease Resistance Gene". Plant Physiology. 110 (4): 1381–1394. doi:10.1104/pp.110.4.1381. PMC 160933 . PMID 12226268.
- ↑ May, M. J.; Hammond-Kosack, K. E.; Jones, J. (1996). "Involvement of Reactive Oxygen Species, Glutathione Metabolism, and Lipid Peroxidation in the Cf-Gene-Dependent Defense Response of Tomato Cotyledons Induced by Race-Specific Elicitors of Cladosporium fulvum". Plant Physiology. 110 (4): 1367–1379. doi:10.1104/pp.110.4.1367. PMC 160932 . PMID 12226267.
- ↑ English, J. J.; Harrison, K.; Jones, J. (1995). "Aberrant Transpositions of Maize Double Ds-Like Elements Usually Involve Ds Ends on Sister Chromatids". The Plant Cell Online. 7 (8): 1235–1247. doi:10.1105/tpc.7.8.1235. PMC 160947 . PMID 12242405.
- ↑ Hammond-Kosack, K. E.; Jones, D. A.; Jones, J. (1994). "Identification of Two Genes Required in Tomato for Full Cf-9-Dependent Resistance to Cladosporium fulvum". The Plant Cell Online. 6 (3): 361–374. doi:10.1105/tpc.6.3.361. PMC 160439 . PMID 12244240.
- ↑ Dangl, J. L.; Jones, J. D. G. (2001). "Plant pathogens and integrated defence responses to infection". Nature. 411 (6839): 826–833. Bibcode:2001Natur.411..826D. doi:10.1038/35081161. PMID 11459065. S2CID 4345575.
- ↑ Foreman, J.; Demidchik, V.; Bothwell, J. H. F.; Mylona, P.; Miedema, H.; Torres, M. A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J. D. G.; Davies, J. M.; Dolan, L. (2003). "Reactive oxygen species produced by NADPH oxidase regulate plant cell growth". Nature . 422 (6930): 442–446. Bibcode:2003Natur.422..442F. doi:10.1038/nature01485. PMID 12660786. S2CID 4328808.
- ↑ Hammond-Kosack, K. E.; Jones, J. D. G. (1997). "Plant Disease Resistance Genes". Annual Review of Plant Physiology and Plant Molecular Biology. 48: 575–607. doi:10.1146/annurev.arplant.48.1.575. PMID 15012275. S2CID 28215144.
- ↑ Zipfel, C.; Robatzek, S.; Navarro, L.; Oakeley, E. J.; Jones, J. D. G.; Felix, G.; Boller, T. (2004). "Bacterial disease resistance in Arabidopsis through flagellin perception". Nature. 428 (6984): 764–767. Bibcode:2004Natur.428..764Z. doi:10.1038/nature02485. PMID 15085136. S2CID 4332562.
- ↑ Jones, Jonathan Dallas George (1980). Repeated DNA sequences in rye (Secale cereale), wheat (Triticum aestivum) and their relatives. jisc.ac.uk (PhD thesis). University of Cambridge. OCLC 53605533. EThOS uk.bl.ethos.258966.
- ↑ Viegas, Jennifer (9 October 2018). "Profile of Jonathan D. G. Jones". Proceedings of the National Academy of Sciences. 115 (41): 10191–10194. Bibcode:2018PNAS..11510191V. doi: 10.1073/pnas.1815072115 . ISSN 0027-8424. PMC 6187187 . PMID 30249645.
- ↑ Jonathan Jones ORCID 0000-0002-4953-261X
- ↑ Jonathan D. G. Jones publications from Europe PubMed Central
- ↑ Jones, J. D. G.; Dangl, J. L. (2006). "The plant immune system". Nature . 444 (7117): 323–9. Bibcode:2006Natur.444..323J. doi: 10.1038/nature05286 . PMID 17108957.
- ↑ Van Der Biezen, E.; Jones, J. D. G. (1998). "Plant disease-resistance proteins and the gene-for-gene concept". Trends in Biochemical Sciences. 23 (12): 454–6. doi:10.1016/S0968-0004(98)01311-5. PMID 9868361.
- ↑ Doward, Jamie (2010). "Scientist leading GM crop test defends links to US biotech giant Monsanto". The Guardian .
- ↑ Smith, Alison Mary; Coupand, George; Dolan, Liam; Harberd, Nicholas; Jones, Jonathan; Martin, Cathie; Sablowski, Robert; Amey, Abigail (2009). Plant Biology . Garland Science. ISBN 978-0815340256.
- ↑ Jones, Jonathan (2010). "Fussy eaters – what's wrong with GM food?". bbc.co.uk. London: BBC News.
![]() This article incorporates text available under the CC BY 4.0 license.
This article incorporates text available under the CC BY 4.0 license.