
Pyrrolysine is an α-amino acid that is used in the biosynthesis of proteins in some methanogenic archaea and bacteria; it is not present in humans. It contains an α-amino group, a carboxylic acid group. Its pyrroline side-chain is similar to that of lysine in being basic and positively charged at neutral pH.

A transferase is any one of a class of enzymes that catalyse the transfer of specific functional groups from one molecule to another. They are involved in hundreds of different biochemical pathways throughout biology, and are integral to some of life's most important processes.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.

Phenylethanolamine N-methyltransferase (PNMT) is an enzyme found primarily in the adrenal medulla that converts norepinephrine (noradrenaline) to epinephrine (adrenaline). It is also expressed in small groups of neurons in the human brain and in selected populations of cardiomyocytes.
Glutamate formimidoyltransferase is a methyltransferase enzyme which uses tetrahydrofolate as part of histidine catabolism. It catalyses two reactions:
In enzymology, a 3'-hydroxy-N-methyl-(S)-coclaurine 4'-O-methyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a 5-methyltetrahydropteroyltriglutamate—homocysteine S-methyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, a protein-glutamate O-methyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a protein-histidine N-methyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a (S)-tetrahydroprotoberberine N-methyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a N-methylalanine dehydrogenase (EC 1.4.1.17) is an enzyme that catalyzes the chemical reaction
In enzymology, a glutamate—methylamine ligase is an enzyme that catalyzes the chemical reaction

The enzyme protein-glutamate methylesterase (EC 3.1.1.61) catalyzes the reaction
In enzymology, a formimidoylglutamate deiminase (EC 3.5.3.13) is an enzyme that catalyzes the chemical reaction
In enzymology, a N-methyl-2-oxoglutaramate hydrolase (EC 3.5.1.36) is an enzyme that catalyzes the chemical reaction
(Methyl-Co methylamine-specific corrinoid protein):coenzyme M methyltransferase is an enzyme with systematic name methylated monomethylamine-specific corrinoid protein:coenzyme M methyltransferase. This enzyme catalyses the following chemical reaction
Methylamine-corrinoid protein Co-methyltransferase is an enzyme with systematic name monomethylamine:5-hydroxybenzimidazolylcobamide Co-methyltransferase. This enzyme catalyses the following chemical reaction
Dimethylamine-corrinoid protein Co-methyltransferase is an enzyme with systematic name dimethylamine:5-hydroxybenzimidazolylcobamide Co-methyltransferase. This enzyme catalyses the following chemical reaction
Trimethylamine-corrinoid protein Co-methyltransferase is an enzyme with systematic name trimethylamine:5-hydroxybenzimidazolylcobamide Co-methyltransferase. This enzyme catalyses the following chemical reaction
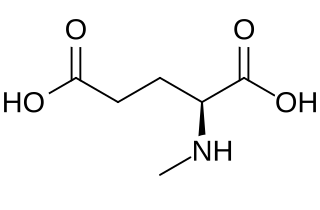
N-Methyl-l-glutamic acid (methylglutamate) is a chemical derivative of glutamic acid in which a methyl group has been added to the amino group. It is an intermediate in methane metabolism. Biosynthetically, it is produced from methylamine and glutamic acid by the enzyme methylamine—glutamate N-methyltransferase. It can also be demethylated by methylglutamate dehydrogenase to regenerate glutamic acid.








