| Formiminotransferase domain, N-terminal subdomain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Formiminotransferase domain of formiminotransferase-cyclodeaminase, homodimer, Sus scrofa | |||||||||
| Identifiers | |||||||||
| Symbol | FTCD_N | ||||||||
| Pfam | PF07837 | ||||||||
| InterPro | IPR012886 | ||||||||
| SCOP2 | 1qd1 / SCOPe / SUPFAM | ||||||||
| |||||||||
| Formiminotransferase domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 the crystal structure of the formiminotransferase domain of formiminotransferase-cyclodeaminase. | |||||||||
| Identifiers | |||||||||
| Symbol | FTCD | ||||||||
| Pfam | PF02971 | ||||||||
| InterPro | IPR013802 | ||||||||
| SCOP2 | 1qd1 / SCOPe / SUPFAM | ||||||||
| |||||||||
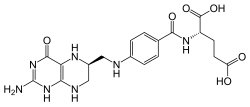
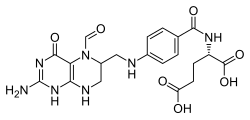
Glutamate formimidoyltransferase is a methyltransferase enzyme which uses tetrahydrofolate as part of histidine catabolism. It catalyses two reactions. In the first, N-formimidoyl-L-glutamate (from histidine) transfers its formimidoyl group to tetrahydrofolate. [1] [2]
Contents
Alternatively, the enzyme can catalyse the transfer of a formyl group from N-formyl-L-glutamic acid. [2]
It is classified under EC 2.1.2.5 and in mammals is found as part of a bifunctional enzyme that also has formimidoyltetrahydrofolate cyclodeaminase activity. [3]