Autolysins are endogenous lytic enzymes that break down the peptidoglycan components of biological cells which enables the separation of daughter cells following cell division. They are involved in cell growth, cell wall metabolism, cell division and separation, as well as peptidoglycan turnover and have similar functions to lysozymes.

Teichoic acids are bacterial copolymers of glycerol phosphate or ribitol phosphate and carbohydrates linked via phosphodiester bonds.

Lysins, also known as endolysins or murein hydrolases, are hydrolytic enzymes produced by bacteriophages in order to cleave the host's cell wall during the final stage of the lytic cycle. Lysins are highly evolved enzymes that are able to target one of the five bonds in peptidoglycan (murein), the main component of bacterial cell walls, which allows the release of progeny virions from the lysed cell. Cell-wall-containing Archaea are also lysed by specialized pseudomurein-cleaving lysins, while most archaeal viruses employ alternative mechanisms. Similarly, not all bacteriophages synthesize lysins: some small single-stranded DNA and RNA phages produce membrane proteins that activate the host's autolytic mechanisms such as autolysins.

In enzymology, an UDP-N-acetylmuramate dehydrogenase (EC 1.3.1.98) is an enzyme that catalyzes the chemical reaction

In enzymology, an alanine racemase is an enzyme that catalyzes the chemical reaction
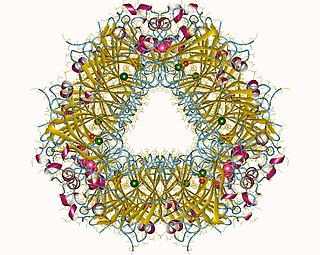
In enzymology, an oxalate decarboxylase (EC 4.1.1.2) is an oxalate degrading enzyme that catalyzes the chemical reaction
In enzymology, a UDP-N-acetylmuramate—L-alanine ligase is an enzyme that catalyzes the chemical reaction
In enzymology, a UDP-N-acetylmuramoyl-L-alanine—D-glutamate ligase is an enzyme that catalyzes the chemical reaction
In enzymology, a UDP-N-acetylmuramoyl-tripeptide—D-alanyl-D-alanine ligase is an enzyme that catalyzes the chemical reaction
In enzymology, a beta-ureidopropionase (EC 3.5.1.6) is an enzyme that catalyzes the chemical reaction
In enzymology, a formimidoylglutamase (EC 3.5.3.8) is an enzyme that catalyzes the chemical reaction
In enzymology, an UDP-N-acetylmuramoylpentapeptide-lysine N6-alanyltransferase (EC 2.3.2.10) is an enzyme that catalyzes the chemical reaction

In enzymology, an UDP-N-acetylglucosamine 1-carboxyvinyltransferase is an enzyme that catalyzes the first committed step in peptidoglycan biosynthesis of bacteria:
In enzymology, a D-amino-acid transaminase is an enzyme that catalyzes the chemical reaction:

Peptidoglycan recognition protein 2(PGLYRP2) is an enzyme, N-acetylmuramoyl-L-alanine amidase (NAMLAA), that hydrolyzes bacterial cell wall peptidoglycan and is encoded by the PGLYRP2 gene.

Peptidoglycan binding domains have a general peptidoglycan binding function and a common core structure consisting of a closed, three-helical bundle with a left-handed twist. It is found at the N or C terminus of a variety of enzymes involved in bacterial cell wall degradation. Examples are:
In molecular biology, glycoside hydrolase family 22 is a family of glycoside hydrolases. EC 3.2.1., which are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycoside hydrolases, based on sequence similarity, has led to the definition of >100 different families. This classification is available on the CAZy web site, and also discussed at CAZypedia, an online encyclopedia of carbohydrate active enzymes.
The bacterial cell wall provides strength and rigidity to counteract internal osmotic pressure, and protection against the environment. The peptidoglycan layer gives the cell wall its strength, and helps maintain the overall shape of the cell. The basic peptidoglycan structure of both Gram-positive and Gram-negative bacteria comprises a sheet of glycan chains connected by short cross-linking polypeptides. Biosynthesis of peptidoglycan is a multi-step process comprising three main stages:
- formation of UDP-N-acetylmuramic acid (UDPMurNAc) from N-acetylglucosamine (GlcNAc).
- addition of a short polypeptide chain to the UDPMurNAc.
- addition of a second GlcNAc to the disaccharide-pentapeptide building block and transport of this unit through the cytoplasmic membrane and incorporation into the growing peptidoglycan layer.
In molecular biology, VanY are protein domains found in enzymes named metallopeptidases. They are vital to bacterial cell wall synthesis and antibiotic resistance.
The LCP family or TagU family of proteins is a conserved family of phosphotransferases that are involved in the attachment of teichoic acid (TA) molecules to gram-positive cell wall or cell membrane. It was initially thought as the LytR component of a LytABC operon encoding autolysins, but the mechanism of regulation was later realized to be the production of TA molecules. It was accordingly renamed TagU.









