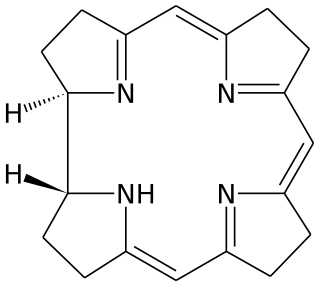
Corrin is a heterocyclic compound. Although not known to exist on its own, the molecule is of interest as the parent macrocycle related to the cofactor and chromophore in vitamin B12. Its name reflects that it is the "core" of vitamin B12 (cobalamins). Compounds with a corrin core are known as "corrins".

Methionine synthase also known as MS, MeSe, MTR is responsible for the regeneration of methionine from homocysteine. In humans it is encoded by the MTR gene (5-methyltetrahydrofolate-homocysteine methyltransferase). Methionine synthase forms part of the S-adenosylmethionine (SAMe) biosynthesis and regeneration cycle, and is the enzyme responsible for linking the cycle to one-carbon metabolism via the folate cycle. There are two primary forms of this enzyme, the Vitamin B12 (cobalamin)-dependent (MetH) and independent (MetE) forms, although minimal core methionine synthases that do not fit cleanly into either category have also been described in some anaerobic bacteria. The two dominant forms of the enzymes appear to be evolutionary independent and rely on considerably different chemical mechanisms. Mammals and other higher eukaryotes express only the cobalamin-dependent form. In contrast, the distribution of the two forms in Archaeplastida (plants and algae) is more complex. Plants exclusively possess the cobalamin-independent form, while algae have either one of the two, depending on species. Many different microorganisms express both the cobalamin-dependent and cobalamin-independent forms.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.
Pseudomonas denitrificans is a Gram-negative aerobic bacterium that performs denitrification. It was first isolated from garden soil in Vienna, Austria. It overproduces cobalamin (vitamin B12), which it uses for methionine synthesis and it has been used for manufacture of the vitamin. Scientists at Rhône-Poulenc Rorer took a genetically engineered strain of the bacteria, in which eight of the cob genes involved in the biosynthesis of the vitamin had been overexpressed, to establish the complete sequence of methylation and other steps in the cobalamin pathway.
In enzymology, a cobalt-factor II C20-methyltransferase is an enzyme that catalyzes the chemical reaction

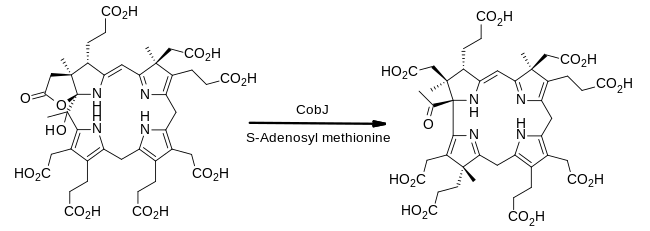
In enzymology, a precorrin-2 C20-methyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a precorrin-4 C11-methyltransferase is an enzyme that catalyzes the chemical reaction
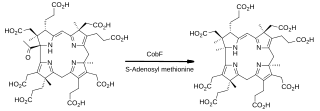
In enzymology, precorrin-6A synthase (deacetylating) (EC 2.1.1.152) is an enzyme that catalyzes the chemical reaction

In enzymology, a precorrin-6Y C5,15-methyltransferase (decarboxylating) (EC 2.1.1.132) is an enzyme that catalyzes the chemical reaction
In enzymology, a precorrin-6A reductase (EC 1.3.1.54) is an enzyme that catalyzes the chemical reaction
In enzymology, a precorrin-3B synthase (EC 1.14.13.83) is an enzyme that catalyzes the chemical reaction

In enzymology, a precorrin-8X methylmutase is an enzyme that catalyzes the chemical reaction

Vitamin B12, also known as cobalamin, is a water-soluble vitamin involved in metabolism. It is one of eight B vitamins. It is required by animals, which use it as a cofactor in DNA synthesis, and in both fatty acid and amino acid metabolism. It is important in the normal functioning of the nervous system via its role in the synthesis of myelin, and in the circulatory system in the maturation of red blood cells in the bone marrow. Plants do not need cobalamin and carry out the reactions with enzymes that are not dependent on it.

In enzymology, a nicotinate-nucleotide-dimethylbenzimidazole phosphoribosyltransferase is an enzyme that catalyzes the chemical reaction

Cobalamin biosynthesis is the process by which bacteria and archea make cobalamin, vitamin B12. Many steps are involved in converting aminolevulinic acid via uroporphyrinogen III and adenosylcobyric acid to the final forms in which it is used by enzymes in both the producing organisms and other species, including humans who acquire it through their diet.

Uroporphyrinogen-III C-methyltransferase, uroporphyrinogen methyltransferase, uroporphyrinogen-III methyltransferase, adenosylmethionine-uroporphyrinogen III methyltransferase, S-adenosyl-L-methionine-dependent uroporphyrinogen III methylase, uroporphyrinogen-III methylase, SirA, CysG, CobA, uroporphyrin-III C-methyltransferase, S-adenosyl-L-methionine:uroporphyrin-III C-methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:uroporphyrinogen-III C-methyltransferase. This enzyme catalyses the following chemical reaction
Cobalt-precorrin-5B (C1)-methyltransferase (EC 2.1.1.195), cobalt-precorrin-6A synthase, CbiD (gene)) is an enzyme with systematic name S-adenosyl-L-methionine:cobalt-precorrin-5B (C1)-methyltransferase. This enzyme catalyses the following chemical reaction
Cobalt-precorrin-7 (C15)-methyltransferase (decarboxylating) (EC 2.1.1.196, CbiT) is an enzyme with systematic name S-adenosyl-L-methionine:precorrin-7 C15-methyltransferase (C12-decarboxylating). This enzyme catalyses the following chemical reaction
Cobalt-precorrin 5A hydrolase (EC 3.7.1.12), CbiG (gene)) is an enzyme with systematic name cobalt-precorrin 5A acylhydrolase. This enzyme catalyses the following chemical reaction

Cobalt is essential to the metabolism of all animals. It is a key constituent of cobalamin, also known as vitamin B12, the primary biological reservoir of cobalt as an ultratrace element. Bacteria in the stomachs of ruminant animals convert cobalt salts into vitamin B12, a compound which can only be produced by bacteria or archaea. A minimal presence of cobalt in soils therefore markedly improves the health of grazing animals, and an uptake of 0.20 mg/kg a day is recommended because they have no other source of vitamin B12.