
Protein biosynthesis is a core biological process, occurring inside cells, balancing the loss of cellular proteins through the production of new proteins. Proteins perform a number of critical functions as enzymes, structural proteins or hormones. Protein synthesis is a very similar process for both prokaryotes and eukaryotes but there are some distinct differences.

Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself or by forming a template for production of proteins. RNA and deoxyribonucleic acid (DNA) are nucleic acids. The nucleic acids constitute one of the four major macromolecules essential for all known forms of life. RNA is assembled as a chain of nucleotides. Cellular organisms use messenger RNA (mRNA) to convey genetic information that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome.

Ribosomes are macromolecular machines, found within all cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.

Ricin ( RY-sin) is a lectin (a carbohydrate-binding protein) and a highly potent toxin produced in the seeds of the castor oil plant, Ricinus communis. The median lethal dose (LD50) of ricin for mice is around 22 micrograms per kilogram of body weight via intraperitoneal injection. Oral exposure to ricin is far less toxic. An estimated lethal oral dose in humans is approximately 1 milligram per kilogram of body weight.
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.
DNA glycosylases are a family of enzymes involved in base excision repair, classified under EC number EC 3.2.2. Base excision repair is the mechanism by which damaged bases in DNA are removed and replaced. DNA glycosylases catalyze the first step of this process. They remove the damaged nitrogenous base while leaving the sugar-phosphate backbone intact, creating an apurinic/apyrimidinic site, commonly referred to as an AP site. This is accomplished by flipping the damaged base out of the double helix followed by cleavage of the N-glycosidic bond.
The peptidyl transferase is an aminoacyltransferase as well as the primary enzymatic function of the ribosome, which forms peptide bonds between adjacent amino acids using tRNAs during the translation process of protein biosynthesis. The substrates for the peptidyl transferase reaction are two tRNA molecules, one bearing the growing peptide chain and the other bearing the amino acid that will be added to the chain. The peptidyl chain and the amino acids are attached to their respective tRNAs via ester bonds to the O atom at the CCA-3' ends of these tRNAs. Peptidyl transferase is an enzyme that catalyzes the addition of an amino acid residue in order to grow the polypeptide chain in protein synthesis. It is located in the large ribosomal subunit, where it catalyzes the peptide bond formation. It is composed entirely of RNA. The alignment between the CCA ends of the ribosome-bound peptidyl tRNA and aminoacyl tRNA in the peptidyl transferase center contribute to its ability to catalyze these reactions. This reaction occurs via nucleophilic displacement. The amino group of the aminoacyl tRNA attacks the terminal carboxyl group of the peptidyl tRNA. Peptidyl transferase activity is carried out by the ribosome. Peptidyl transferase activity is not mediated by any ribosomal proteins but by ribosomal RNA (rRNA), a ribozyme. Ribozymes are the only enzymes which are not made up of proteins, but ribonucleotides. All other enzymes are made up of proteins. This RNA relic is the most significant piece of evidence supporting the RNA World hypothesis.
Protein metabolism denotes the various biochemical processes responsible for the synthesis of proteins and amino acids (anabolism), and the breakdown of proteins by catabolism.
Saporin is a protein that is useful in biological research applications, especially studies of behavior. Saporins are so-called ribosome inactivating proteins (RIPs), due to its N-glycosidase activity, from the seeds of Saponaria officinalis. It was first described by Fiorenzo Stirpe and his colleagues in 1983 in an article that illustrated the unusual stability of the protein.

EF-G is a prokaryotic elongation factor involved in protein translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome.
Beetin is a ribosome-inactivating protein found in the leaves of sugar beets, Beta vulgaris L, specifically attacking plant ribosomes. Sugar beet, beetins, that have been isolated meet all the criteria to be classified as single chain ribosome inactivating proteins that are highly toxic to mammalian ribosomes but non-toxic to intact cultured mammalian cells. Beetin expression occurs when there is a viral infection of the plant. The different levels of glycosylation of the same polypeptide chain result in the two forms of beetin. Beetin exhibits these two primary forms with apparent Mr values of 27 000 (BE27) and 29 000 (BE29) along with possessing glycan chains. Beetins are a type-I (single-chain) proteins with N-glycoside activity. Since it has been discovered that beetin is mostly concentrated in the intercellular fluid, its presence in the remaining parts of the leaf may be below the limit of detection rather than being nonexistent. The expression of beetin is only found in mature plants, but is present in all developing stages.

A protein synthesis inhibitor is a compound that stops or slows the growth or proliferation of cells by disrupting the processes that lead directly to the generation of new proteins.

A ribosome-inactivating protein (RIP) is a protein synthesis inhibitor that acts at the eukaryotic ribosome. This protein family describes a large family of such proteins that work by acting as rRNA N-glycosylase. They inactivate 60S ribosomal subunits by an N-glycosidic cleavage, which releases a specific adenine base from the sugar-phosphate backbone of 28S rRNA. RIPs exist in bacteria and plants.

Toxalbumins are toxic plant proteins that disable ribosomes and thereby inhibit protein synthesis, producing severe cytotoxic effects in multiple organ systems. They are dimers held together by a disulfide bond and comprise a lectin part which binds to the cell membrane and enables the toxin part to gain access to the cell contents. Toxalbumins are similar in structure to AB toxins found in cholera, tetanus, diphtheria, botulinum and others; and their physiological and toxic properties are similar to those of viperine snake venom.

The FPG IleRS zinc finger domain represents a zinc finger domain found at the C-terminal in both DNA glycosylase/AP lyase enzymes and in isoleucyl tRNA synthetase. In these two types of enzymes, the C-terminal domain forms a zinc finger.
rRNA endonuclease is an enzyme that catalyses the hydrolysis of the phosphodiester linkage between guanosine and adenosine residues at one specific position in the 28S rRNA of rat ribosomes. This enzyme also acts on bacterial rRNA.
Deoxyribodipyrimidine endonucleosidase is an enzyme with systematic name deoxy-D-ribocyclobutadipyrimidine polynucleotidodeoxyribohydrolase. This enzyme catalyses the following chemical reaction
DNA-formamidopyrimidine glycosylase is an enzyme with systematic name DNA glycohydrolase . FPG is a base excision repair enzyme which recognizes and removes a wide range of oxidized purines from correspondingly damaged DNA. It was discovered by Zimbabwean scientist Christopher J. Chetsanga in 1975.
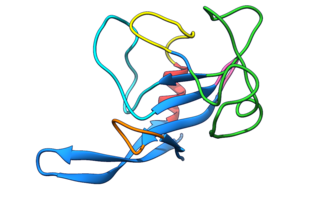
Fungal ribotoxins are a group of extracellular ribonucleases (RNases) secreted by fungi. Their most notable characteristic is their extraordinary specificity. They inactivate ribosomes by cutting a single phosphodiester bond of the rRNA that is found in a universally conserved sequence. This cleavage leads to cell death by apoptosis. However, since they are extracellular proteins, they must first enter the cells that constitute their target to exert their cytotoxic action. This entry constitutes the rate-determining step of their action.
Modeccin is a toxic lectin, a group of glycoproteins capable of binding specifically to sugar moieties. Different toxic lectins are present in seeds of different origin. Modeccin is found in the roots of the African plant Adenia digitata. These roots are often mistaken for edible roots, which has led to some cases of intoxication. Sometimes the fruit is eaten, or a root extract is drunk as a manner of suicide.










