Ferredoxins are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium Clostridium pasteurianum.

Sulfate-reducing microorganisms (SRM) or sulfate-reducing prokaryotes (SRP) are a group composed of sulfate-reducing bacteria (SRB) and sulfate-reducing archaea (SRA), both of which can perform anaerobic respiration utilizing sulfate (SO2−
4) as terminal electron acceptor, reducing it to hydrogen sulfide (H2S). Therefore, these sulfidogenic microorganisms "breathe" sulfate rather than molecular oxygen (O2), which is the terminal electron acceptor reduced to water (H2O) in aerobic respiration.
Iron–sulfur proteins are proteins characterized by the presence of iron–sulfur clusters containing sulfide-linked di-, tri-, and tetrairon centers in variable oxidation states. Iron–sulfur clusters are found in a variety of metalloproteins, such as the ferredoxins, as well as NADH dehydrogenase, hydrogenases, coenzyme Q – cytochrome c reductase, succinate – coenzyme Q reductase and nitrogenase. Iron–sulfur clusters are best known for their role in the oxidation-reduction reactions of electron transport in mitochondria and chloroplasts. Both Complex I and Complex II of oxidative phosphorylation have multiple Fe–S clusters. They have many other functions including catalysis as illustrated by aconitase, generation of radicals as illustrated by SAM-dependent enzymes, and as sulfur donors in the biosynthesis of lipoic acid and biotin. Additionally, some Fe–S proteins regulate gene expression. Fe–S proteins are vulnerable to attack by biogenic nitric oxide, forming dinitrosyl iron complexes. In most Fe–S proteins, the terminal ligands on Fe are thiolate, but exceptions exist.
Any enzyme system that includes cytochrome P450 protein or domain can be called a P450-containing system.

Sulfur assimilation is the process by which living organisms incorporate sulfur into their biological molecules. In plants, sulfate is absorbed by the roots and then be transported to the chloroplasts by the transipration stream where the sulfur are reduced to sulfide with the help of a series of enzymatic reactions. Furthermore, the reduced sulfur is incorporated into cysteine, an amino acid that is a precursor to many other sulfur-containing compounds. In animals, sulfur assimilation occurs primarily through the diet, as animals cannot produce sulfur-containing compounds directly. Sulfur is incorporated into amino acids such as cysteine and methionine, which are used to build proteins and other important molecules. Besides, With the rapid development of economy, the increase emission of sulfur results in environmental issues, such as acid rain and hydrogen sulfilde.
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.
In enzymology, a ferredoxin-NADP+ reductase (EC 1.18.1.2) abbreviated FNR, is an enzyme that catalyzes the chemical reaction
In enzymology, a sulfur oxygenase/reductase (EC 1.13.11.55) is an enzyme that catalyzes the chemical reaction
Adenylyl-sulfate reductase is an enzyme that catalyzes the chemical reaction of the reduction of adenylyl-sulfate/adenosine-5'-phosphosulfate (APS) to sulfite through the use of an electron donor cofactor. The products of the reaction are AMP and sulfite, as well as an oxidized electron donor cofactor.
In enzymology, a ferredoxin—nitrite reductase (EC 1.7.7.1) is an enzyme that catalyzes the chemical reaction

Sulfite reductases (EC 1.8.99.1) are enzymes that participate in sulfur metabolism. They catalyze the reduction of sulfite to hydrogen sulfide and water. Electrons for the reaction are provided by a dissociable molecule of either NADPH, bound flavins, or ferredoxins.
Sulfite reductase (NADPH) (EC 1.8.1.2, sulfite (reduced nicotinamide adenine dinucleotide phosphate) reductase, NADPH-sulfite reductase, NADPH-dependent sulfite reductase, H2S-NADP oxidoreductase, sulfite reductase (NADPH2)) is an enzyme with systematic name hydrogen-sulfide:NADP+ oxidoreductase. This enzyme catalises the following chemical reaction
In enzymology, a thiosulfate-dithiol sulfurtransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a thiosulfate-thiol sulfurtransferase is an enzyme that catalyzes the chemical reaction
Sulfur is metabolized by all organisms, from bacteria and archaea to plants and animals. Sulfur can have an oxidation state from -2 to +6 and is reduced or oxidized by a diverse range of organisms. The element is present in proteins, sulfate esters of polysaccharides, steroids, phenols, and sulfur-containing coenzymes.
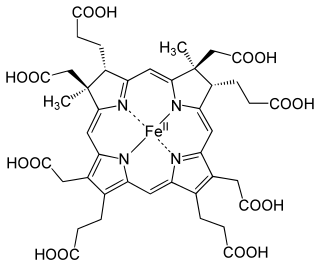
Siroheme is a heme-like prosthetic group at the active sites of some enzymes to accomplish the six-electron reduction of sulfur and nitrogen. It is a cofactor at the active site of sulfite reductase, which plays a major role in sulfur assimilation pathway, converting sulfite into sulfide, which can be incorporated into the organic compound homocysteine.

Dissimilatory sulfate reduction is a form of anaerobic respiration that uses sulfate as the terminal electron acceptor to produce hydrogen sulfide. This metabolism is found in some types of bacteria and archaea which are often termed sulfate-reducing organisms. The term "dissimilatory" is used when hydrogen sulfide is produced in an anaerobic respiration process. By contrast, the term "assimilatory" would be used in relation to the biosynthesis of organosulfur compounds, even though hydrogen sulfide may be an intermediate.
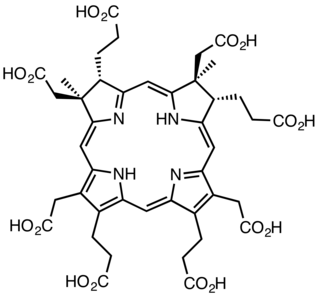
Sirohydrochlorin is a tetrapyrrole macrocyclic metabolic intermediate in the biosynthesis of sirohaem, the iron-containing prosthetic group in sulfite reductase enzymes. It is also the biosynthetic precursor to cofactor F430, an enzyme which catalyzes the release of methane in the final step of methanogenesis.
Dissimilatory sulfite reductase is an enzyme that participates in sulfur metabolism in dissimilatory sulfate reduction.









