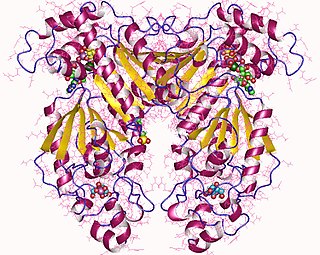
Phosphofructokinase-2 (6-phosphofructo-2-kinase, PFK-2) or fructose bisphosphatase-2 (FBPase-2), is an enzyme indirectly responsible for regulating the rates of glycolysis and gluconeogenesis in cells. It catalyzes formation and degradation of a significant allosteric regulator, fructose-2,6-bisphosphate (Fru-2,6-P2) from substrate fructose-6-phosphate. Fru-2,6-P2 contributes to the rate-determining step of glycolysis as it activates enzyme phosphofructokinase 1 in the glycolysis pathway, and inhibits fructose-1,6-bisphosphatase 1 in gluconeogenesis. Since Fru-2,6-P2 differentially regulates glycolysis and gluconeogenesis, it can act as a key signal to switch between the opposing pathways. Because PFK-2 produces Fru-2,6-P2 in response to hormonal signaling, metabolism can be more sensitively and efficiently controlled to align with the organism's glycolytic needs. This enzyme participates in fructose and mannose metabolism. The enzyme is important in the regulation of hepatic carbohydrate metabolism and is found in greatest quantities in the liver, kidney and heart. In mammals, several genes often encode different isoforms, each of which differs in its tissue distribution and enzymatic activity. The family described here bears a resemblance to the ATP-driven phospho-fructokinases, however, they share little sequence similarity, although a few residues seem key to their interaction with fructose 6-phosphate.

Phosphoglycerate mutase (PGM) is any enzyme that catalyzes step 8 of glycolysis - the internal transfer of a phosphate group from C-3 to C-2 which results in the conversion of 3-phosphoglycerate (3PG) to 2-phosphoglycerate (2PG) through a 2,3-bisphosphoglycerate intermediate. These enzymes are categorized into the two distinct classes of either cofactor-dependent (dPGM) or cofactor-independent (iPGM). The dPGM enzyme is composed of approximately 250 amino acids and is found in all vertebrates as well as in some invertebrates, fungi, and bacteria. The iPGM class is found in all plants and algae as well as in some invertebrate, fungi, and Gram-positive bacteria. This class of PGM enzyme shares the same superfamily as alkaline phosphatase.

Methylmalonyl-CoA mutase (EC 5.4.99.2, MCM), mitochondrial, also known as methylmalonyl-CoA isomerase, is a protein that in humans is encoded by the MUT gene. This vitamin B12-dependent enzyme catalyzes the isomerization of methylmalonyl-CoA to succinyl-CoA in humans. Mutations in MUT gene may lead to various types of methylmalonic aciduria.

Propionyl-CoA carboxylase (EC 6.4.1.3, PCC) catalyses the carboxylation reaction of propionyl-CoA in the mitochondrial matrix. PCC has been classified both as a ligase and a lyase. The enzyme is biotin-dependent. The product of the reaction is (S)-methylmalonyl CoA.

Steroid 11β-hydroxylase, also known as steroid 11β-monooxygenase, is a steroid hydroxylase found in the zona glomerulosa and zona fasciculata of the adrenal cortex. Named officially the cytochrome P450 11B1, mitochondrial, it is a protein that in humans is encoded by the CYP11B1 gene. The enzyme is involved in the biosynthesis of adrenal corticosteroids by catalyzing the addition of hydroxyl groups during oxidation reactions.
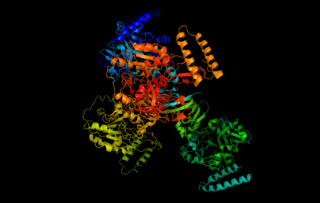
UTP—glucose-1-phosphate uridylyltransferase also known as glucose-1-phosphate uridylyltransferase is an enzyme involved in carbohydrate metabolism. It synthesizes UDP-glucose from glucose-1-phosphate and UTP; i.e.,
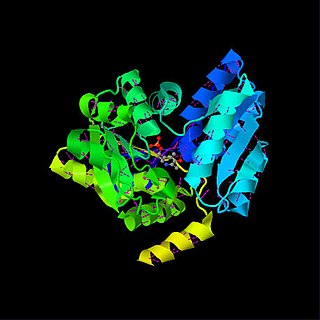
Serine dehydratase or L-serine ammonia lyase (SDH) is in the β-family of pyridoxal phosphate-dependent (PLP) enzymes. SDH is found widely in nature, but its structural and properties vary among species. SDH is found in yeast, bacteria, and the cytoplasm of mammalian hepatocytes. SDH catalyzes is the deamination of L-serine to yield pyruvate, with the release of ammonia.

In enzymology, a phosphoenolpyruvate mutase is an enzyme that catalyzes the chemical reaction

In enzymology, a galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a glycoprotein 2-beta-D-xylosyltransferase (EC 2.4.2.38) is an enzyme that catalyzes the chemical reaction

In enzymology, a sucrose synthase is an enzyme that catalyzes the chemical reaction

3-beta-glucuronosyltransferase 1 (B3GAT1) is an enzyme that in humans is encoded by the B3GAT1 gene, whose enzymatic activity creates the CD57 epitope on other cell surface proteins. In immunology, the CD57 antigen is also known as HNK1 or LEU7. It is expressed as a carbohydrate epitope that contains a sulfoglucuronyl residue in several adhesion molecules of the nervous system.

UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 1 is an enzyme that in humans is encoded by the B3GALNT1 gene.

Beta-1,4 N-acetylgalactosaminyltransferase 1 is an enzyme that in humans is encoded by the B4GALNT1 gene.
UDP-4-amino-4-deoxy-L-arabinose formyltransferase is an enzyme with systematic name 10-formyltetrahydrofolate:UDP-4-amino-4-deoxy-beta-L-arabinose N-formyltransferase. This enzyme catalyses the following chemical reaction
N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase is an enzyme with systematic name UDP-N-acetyl-D-galactosamine:N-acetyl-beta-D-glucosaminyl-group 4-beta-N-acetylgalactosaminyltransferase. This enzyme catalyses the following chemical reaction
The alpha-D-phosphohexomutases are a large superfamily of enzymes, with members in all three domains of life. Enzymes from this superfamily are ubiquitous in organisms from E. Coli to humans, and catalyze a phosphoryl transfer reaction on a phosphosugar substrate. Four well studied subgroups in the superfamily are:
- Phosphoglucomutase (PGM)
- Phosphoglucomutase/Phosphomannomutase (PGM/PMM)
- Phosphoglucosamine mutase (PNGM)
- Phosphoaceytlglucosamine mutase (PAGM)
UDP-4-amino-4-deoxy-L-arabinose aminotransferase is an enzyme with systematic name UDP-4-amino-4-deoxy-beta-L-arabinose:2-oxoglutarate aminotransferase. This enzyme catalyses the following chemical reaction
UDP-4-amino-4,6-dideoxy-N-acetyl-beta-L-altrosamine transaminase is an enzyme with systematic name UDP-4-amino-4,6-dideoxy-N-acetyl-beta-L-altrosamine:2-oxoglutarate aminotransferase. This enzyme catalyses the following chemical reaction
Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase is an enzyme with systematic name UDP-4-amino-4-deoxy-alpha-L-arabinose:ditrans,octacis-undecaprenyl phosphate 4-amino-4-deoxy-alpha-L-arabinosyltransferase. This enzyme catalyses the following chemical reaction













