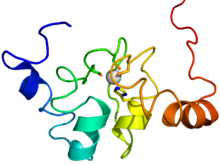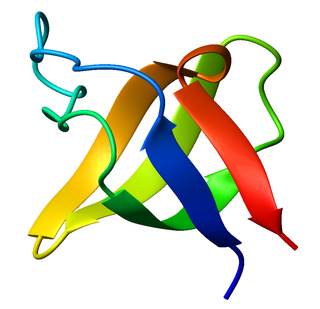
The SRC Homology 3 Domain is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src. It has also been identified in several other protein families such as: PI3 Kinase, Ras GTPase-activating protein, CDC24 and cdc25. SH3 domains are found in proteins of signaling pathways regulating the cytoskeleton, the Ras protein, and the Src kinase and many others. The SH3 proteins interact with adaptor proteins and tyrosine kinases. Interacting with tyrosine kinases, SH3 proteins usually bind far away from the active site. Approximately 300 SH3 domains are found in proteins encoded in the human genome. In addition to that, the SH3 domain was responsible for controlling protein-protein interactions in the signal transduction pathways and regulating the interactions of proteins involved in the cytoplasmic signaling.

14-3-3 proteins are a family of conserved regulatory molecules that are expressed in all eukaryotic cells. 14-3-3 proteins have the ability to bind a multitude of functionally diverse signaling proteins, including kinases, phosphatases, and transmembrane receptors. More than 200 signaling proteins have been reported as 14-3-3 ligands.

The ankyrin repeat is a 33-residue motif in proteins consisting of two alpha helices separated by loops, first discovered in signaling proteins in yeast Cdc10 and Drosophila Notch. Domains consisting of ankyrin tandem repeats mediate protein–protein interactions and are among the most common structural motifs in known proteins. They appear in bacterial, archaeal, and eukaryotic proteins, but are far more common in eukaryotes. Ankyrin repeat proteins, though absent in most viruses, are common among poxviruses. Most proteins that contain the motif have four to six repeats, although its namesake ankyrin contains 24, and the largest known number of repeats is 34, predicted in a protein expressed by Giardia lamblia.

Survivin, also called baculoviral inhibitor of apoptosis repeat-containing 5 or BIRC5, is a protein that, in humans, is encoded by the BIRC5 gene.
Inhibitors of apoptosis are a group of proteins that mainly act on the intrinsic pathway that block programmed cell death, which can frequently lead to cancer or other effects for the cell if mutated or improperly regulated. Many of these inhibitors act to block caspases, a family of cysteine proteases that play an integral role in apoptosis. Some of these inhibitors include the Bcl-2 family, viral inhibitor crmA, and IAP's.

X-linked inhibitor of apoptosis protein (XIAP), also known as inhibitor of apoptosis protein 3 (IAP3) and baculoviral IAP repeat-containing protein 4 (BIRC4), is a protein that stops apoptotic cell death. In humans, this protein (XIAP) is produced by a gene named XIAP gene located on the X chromosome.
Nerve tissue is a biological molecule related to the function and maintenance of normal nervous tissue. An example would include, for example, the generation of myelin which insulates and protects nerves. These are typically calcium-binding proteins.

Baculoviral IAP repeat-containing protein3 is a protein that in humans is encoded by the BIRC3 gene.

Baculoviral IAP repeat-containing protein 2 is a protein that in humans is encoded by the BIRC2 gene.

Diablo homolog (DIABLO) is a mitochondrial protein that in humans is encoded by the DIABLO gene on chromosome 12. DIABLO is also referred to as second mitochondria-derived activator of caspases or SMAC. This protein binds inhibitor of apoptosis proteins (IAPs), thus freeing caspases to activate apoptosis. Due to its proapoptotic function, SMAC is implicated in a broad spectrum of tumors, and small molecule SMAC mimetics have been developed to enhance current cancer treatments.

Baculoviral IAP repeat-containing protein 7 is a protein that in humans is encoded by the BIRC7 gene.

Baculoviral IAP repeat-containing protein 6 is a protein that in humans is encoded by the BIRC6 gene.

The WD40 repeat is a short structural motif of approximately 40 amino acids, often terminating in a tryptophan-aspartic acid (W-D) dipeptide. Tandem copies of these repeats typically fold together to form a type of circular solenoid protein domain called the WD40 domain.
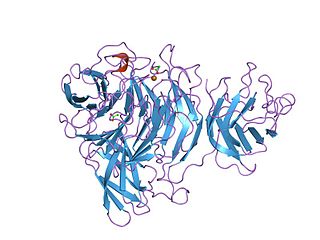
The Kelch motif is a region of protein sequence found widely in proteins from bacteria and eukaryotes. This sequence motif is composed of about 50 amino acid residues which form a structure of a four stranded beta-sheet "blade". This sequence motif is found in between five and eight tandem copies per protein which fold together to form a larger circular solenoid structure called a beta-propeller domain.
The NACHT domain is an evolutionarily conserved protein domain. This NTPase domain is found in apoptosis proteins as well as those involved in MHC transcription. Its name reflects some of the proteins that contain it: NAIP, CIITA, HET-E and TEP1.

BRCA1 C Terminus (BRCT) domain is a family of evolutionarily related proteins. It is named after the C-terminal domain of BRCA1, a DNA-repair protein that serves as a marker of breast cancer susceptibility.
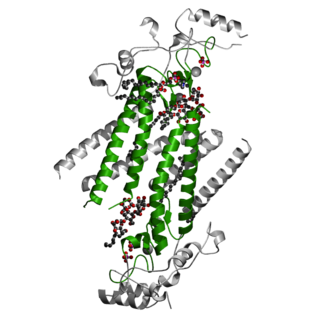
In molecular biology the DHHC domain is a protein domain that acts as an enzyme, which adds a palmitoyl chemical group to proteins in order to anchor them to cell membranes. The DHHC domain was discovered in 1999 and named after a conserved sequence motif found in its protein sequence. Roth and colleagues showed that the yeast Akr1p protein could palmitoylate Yck2p in vitro and inferred that the DHHC domain defined a large family of palmitoyltransferases. In mammals twenty three members of this family have been identified and their substrate specificities investigated. Some members of the family such as ZDHHC3 and ZDHHC7 enhance palmitoylation of proteins such as PSD-95, SNAP-25, GAP43, Gαs. Others such as ZDHHC9 showed specificity only toward the H-Ras protein. However, a recent study questions the involvement of classical enzyme-substrate recognition and specificity in the palmitoylation reaction. Several members of the family have been implicated in human diseases.

In molecular biology the MYND-type zinc finger domain is a conserved protein domain. The MYND domain is present in a large group of proteins that includes RP-8 (PDCD2), Nervy, and predicted proteins from Drosophila, mammals, Caenorhabditis elegans, yeast, and plants. The MYND domain consists of a cluster of cysteine and histidine residues, arranged with an invariant spacing to form a potential zinc-binding motif. Mutating conserved cysteine residues in the DEAF-1 MYND domain does not abolish DNA binding, which suggests that the MYND domain might be involved in protein-protein interactions. Indeed, the MYND domain of ETO/MTG8 interacts directly with the N-CoR and SMRT co-repressors. Aberrant recruitment of co-repressor complexes and inappropriate transcriptional repression is believed to be a general mechanism of leukemogenesis caused by the t(8;21) translocations that fuse ETO with the acute myelogenous leukemia 1 (AML1) protein. ETO has been shown to be a co-repressor recruited by the promyelocytic leukemia zinc finger (PLZF) protein. A divergent MYND domain present in the adenovirus E1A binding protein BS69 was also shown to interact with N-CoR and mediate transcriptional repression. The current evidence suggests that the MYND motif in mammalian proteins constitutes a protein-protein interaction domain that functions as a co-repressor-recruiting interface.
WH1 domain is an evolutionary conserved protein domain found on WASP proteins, which are often involved in actin polymerization.
cIAP1 is the abbreviation for a human protein, cellular inhibitor of apoptosis protein-1. It belongs to the IAP family of proteins and therefore contains at least one BIR domain. cIAP1 is a multi-functional protein which can be found in the cytoplasm of cells and in the nucleus of tumor cells. Its function in this particular case is yet to be understood. However, it is well known that this protein has a big influence in the growth of diverse cancers. cIAP1 is involved in the development process of osteosarcoma and gastric cancer among others.