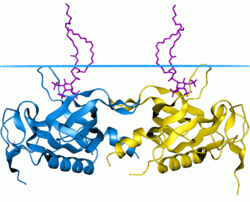
Phosphoinositide phospholipase C is a family of eukaryotic intracellular enzymes that play an important role in signal transduction processes. These enzymes belong to a larger superfamily of Phospholipase C. Other families of phospholipase C enzymes have been identified in bacteria and trypanosomes. Phospholipases C are phosphodiesterases.
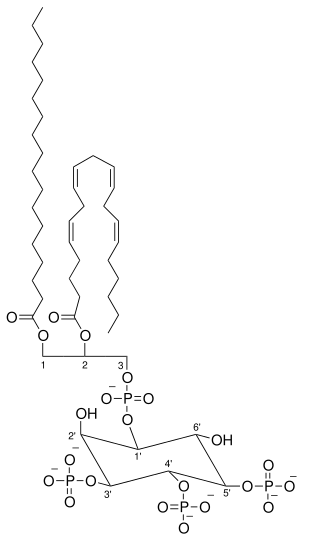
Phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)P3), abbreviated PIP3, is the product of the class I phosphoinositide 3-kinases' (PI 3-kinases) phosphorylation of phosphatidylinositol (4,5)-bisphosphate (PIP2). It is a phospholipid that resides on the plasma membrane.
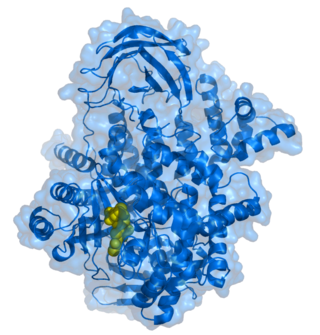
Phosphoinositide 3-kinases (PI3Ks), also called phosphatidylinositol 3-kinases, are a family of enzymes involved in cellular functions such as cell growth, proliferation, differentiation, motility, survival and intracellular trafficking, which in turn are involved in cancer.
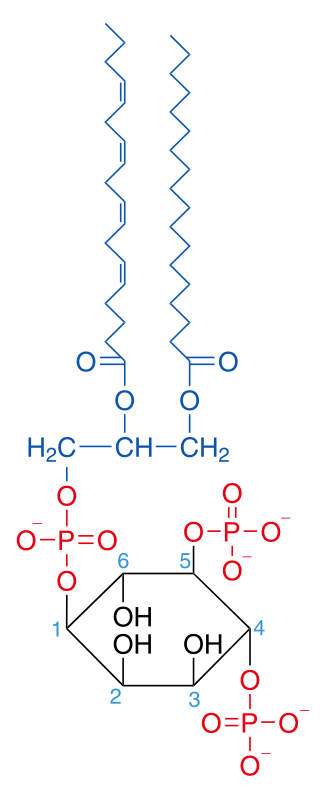
Phosphatidylinositol 4,5-bisphosphate or PtdIns(4,5)P2, also known simply as PIP2 or PI(4,5)P2, is a minor phospholipid component of cell membranes. PtdIns(4,5)P2 is enriched at the plasma membrane where it is a substrate for a number of important signaling proteins. PIP2 also forms lipid clusters that sort proteins.

Phosphatidylinositol 3-phosphate (PtdIns3P) is a phospholipid found in cell membranes that helps to recruit a range of proteins, many of which are involved in protein trafficking, to the membranes. It is the product of both the class II and III phosphoinositide 3-kinases activity on phosphatidylinositol.

Phosphatidylinositol (3,4)-bisphosphate is a minor phospholipid component of cell membranes, yet an important second messenger. The generation of PtdIns(3,4)P2 at the plasma membrane activates a number of important cell signaling pathways.

Phosphatidylinositol 3,5-bisphosphate is one of the seven phosphoinositides found in eukaryotic cell membranes. In quiescent cells, the PtdIns(3,5)P2 levels, typically quantified by HPLC, are the lowest amongst the constitutively present phosphoinositides. They are approximately 3 to 5-fold lower as compared to PtdIns3P and PtdIns5P levels, and more than 100-fold lower than the abundant PtdIns4P and PtdIns(4,5)P2. PtdIns(3,5)P2 was first reported to occur in mouse fibroblasts and budding yeast S. cerevisiae in 1997. In S. cerevisiae PtdIns(3,5)P2 levels increase dramatically during hyperosmotic shock. The response to hyperosmotic challenge is not conserved in most tested mammalian cells except for differentiated 3T3L1 adipocytes.
Phospholipase D (EC 3.1.4.4, lipophosphodiesterase II, lecithinase D, choline phosphatase, PLD; systematic name phosphatidylcholine phosphatidohydrolase) is an enzyme of the phospholipase superfamily that catalyses the following reaction

Phosphatidylinositol 3-kinase regulatory subunit alpha is an enzyme that in humans is encoded by the PIK3R1 gene.
In enzymology, a ceramide kinase, also abbreviated as CERK, is an enzyme that catalyzes the chemical reaction:

Src homology 2 (SH2) domain containing inositol polyphosphate 5-phosphatase 1(SHIP1) is an enzyme with phosphatase activity. SHIP1 is structured by multiple domain and is encoded by the INPP5D gene in humans. SHIP1 is expressed predominantly by hematopoietic cells but also, for example, by osteoblasts and endothelial cells. This phosphatase is important for the regulation of cellular activation. Not only catalytic but also adaptor activities of this protein are involved in this process. Its movement from the cytosol to the cytoplasmic membrane, where predominantly performs its function, is mediated by tyrosine phosphorylation of the intracellular chains of cell surface receptors that SHIP1 binds. Insufficient regulation of SHIP1 leads to different pathologies.

Phospholipase C (PLC) is a class of membrane-associated enzymes that cleave phospholipids just before the phosphate group (see figure). It is most commonly taken to be synonymous with the human forms of this enzyme, which play an important role in eukaryotic cell physiology, in particular signal transduction pathways. Phospholipase C's role in signal transduction is its cleavage of phosphatidylinositol 4,5-bisphosphate (PIP2) into diacyl glycerol (DAG) and inositol 1,4,5-trisphosphate (IP3), which serve as second messengers. Activators of each PLC vary, but typically include heterotrimeric G protein subunits, protein tyrosine kinases, small G proteins, Ca2+, and phospholipids.

Phosphatidylinositol 4-kinase beta is an enzyme that in humans is encoded by the PI4KB gene.

Collagen type IV alpha-3-binding protein, also known as ceramide transfer protein (CERT) or StAR-related lipid transfer protein 11 (STARD11) is a protein that in humans is encoded by the COL4A3BP gene. The protein contains a pleckstrin homology domain at its amino terminus and a START domain towards the end of the molecule. It is a member of the StarD2 subfamily of START domain proteins.

Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3-phosphoinositide is a protein that in humans is encoded by the DAPP1 gene.

Ras GTPase-activating protein 3 is an enzyme that in humans is encoded by the RASA3 gene.

Oxysterol-binding protein 1 is a protein that in humans is encoded by the OSBP gene.

Pleckstrin homology domain-containing family A member 1 is a protein that in humans is encoded by the PLEKHA1 gene.
The Akt signaling pathway or PI3K-Akt signaling pathway is a signal transduction pathway that promotes survival and growth in response to extracellular signals. Key proteins involved are PI3K and Akt.

In the field of biochemistry, PDPK1 refers to the protein 3-phosphoinositide-dependent protein kinase-1, an enzyme which is encoded by the PDPK1 gene in humans. It is implicated in the development and progression of melanomas.