Endocytosis is a cellular process in which substances are brought into the cell. The material to be internalized is surrounded by an area of cell membrane, which then buds off inside the cell to form a vesicle containing the ingested materials. Endocytosis includes pinocytosis and phagocytosis. It is a form of active transport.
GTPases are a large family of hydrolase enzymes that bind to the nucleotide guanosine triphosphate (GTP) and hydrolyze it to guanosine diphosphate (GDP). The GTP binding and hydrolysis takes place in the highly conserved P-loop "G domain", a protein domain common to many GTPases.

Clathrin is a protein that plays a role in the formation of coated vesicles. Clathrin was first isolated by Barbara Pearse in 1976. It forms a triskelion shape composed of three clathrin heavy chains and three light chains. When the triskelia interact they form a polyhedral lattice that surrounds the vesicle. The protein's name refers to this lattice structure, deriving from Latin clathri meaning lattice. Barbara Pearse named the protein clathrin at the suggestion of Graeme Mitchison, selecting it from three possible options. Coat-proteins, like clathrin, are used to build small vesicles in order to transport molecules within cells. The endocytosis and exocytosis of vesicles allows cells to communicate, to transfer nutrients, to import signaling receptors, to mediate an immune response after sampling the extracellular world, and to clean up the cell debris left by tissue inflammation. The endocytic pathway can be hijacked by viruses and other pathogens in order to gain entry to the cell during infection.
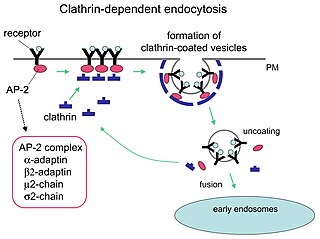
Receptor-mediated endocytosis (RME), also called clathrin-mediated endocytosis, is a process by which cells absorb metabolites, hormones, proteins – and in some cases viruses – by the inward budding of the plasma membrane (invagination). This process forms vesicles containing the absorbed substances and is strictly mediated by receptors on the surface of the cell. Only the receptor-specific substances can enter the cell through this process.

Pleckstrin homology domain or (PHIP) is a protein domain of approximately 120 amino acids that occurs in a wide range of proteins involved in intracellular signaling or as constituents of the cytoskeleton.
Phospholipase D (EC 3.1.4.4, lipophosphodiesterase II, lecithinase D, choline phosphatase, PLD; systematic name phosphatidylcholine phosphatidohydrolase) is an anesthetic sensitive and mechanosensitive enzyme of the phospholipase superfamily that catalyses the following reaction
In molecular biology, BAR domains are highly conserved protein dimerisation domains that occur in many proteins involved in membrane dynamics in a cell. The BAR domain is banana-shaped and binds to membrane via its concave face. It is capable of sensing membrane curvature by binding preferentially to curved membranes. BAR domains are named after three proteins that they are found in: Bin, Amphiphysin and Rvs.

Amphiphysin is a protein that in humans is encoded by the AMPH gene.

Guanine nucleotide exchange factors (GEFs) are proteins or protein domains that activate monomeric GTPases by stimulating the release of guanosine diphosphate (GDP) to allow binding of guanosine triphosphate (GTP). A variety of unrelated structural domains have been shown to exhibit guanine nucleotide exchange activity. Some GEFs can activate multiple GTPases while others are specific to a single GTPase.
Synaptojanin is a protein involved in vesicle uncoating in neurons. This is an important regulatory lipid phosphatase. It dephosphorylates the D-5 position phosphate from phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and phosphatidylinositol (4,5)-bisphosphate (PIP2). It belongs to family of 5-phosphatases, which are structurally unrelated to D-3 inositol phosphatases like PTEN. Other members of the family of 5'phosphoinositide phosphatases include OCRL, SHIP1, SHIP2, INPP5J, INPP5E, INPP5B, INPP5A and SKIP.
Dynamin-2 is a protein that in humans is encoded by the DNM2 gene.

Intersectin-1 is a protein that, in humans, is encoded by the ITSN1 gene.

Dynamin-1 is a protein that in humans is encoded by the DNM1 gene.

Dynamin-1-like protein is a GTPase that regulates mitochondrial fission. In humans, dynamin-1-like protein, which is typically referred to as dynamin-related protein 1 (Drp1), is encoded by the DNM1L gene and is part of the dynamin superfamily (DSP) family of proteins.
The EHD protein family is a relatively small group of proteins which have been shown to play a role in several physiological functions, the most notable being the regulation of endocytotic vesicles. This family is recognized by its highly conserved EH domain, a structural motif that has been shown to facilitate specificity and interaction between protein and ligand. The four mammalian EHD proteins that have been classified are: EHD1, EHD2, EHD3, and EHD4.
Bulk endocytosis refers to a form of endocytosis of synaptic vesicles at nerve terminals. In bulk endocytosis, compared to clathrin-mediated endocytosis, a larger area of presynaptic plasma membrane is internalised as cisternae or endosomes from which multiple synaptic vesicles can subsequently bud off. Bulk endocytosis is activated specifically during intense stimulation, such as during high-frequency trains of action potentials or in response to membrane depolarization by high extracellular concentrations of potassium.
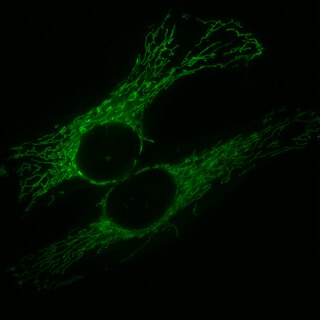
Mitochondrial fission is the process by which mitochondria divide or segregate into two separate mitochondrial organelles. Mitochondrial fission is counteracted by mitochondrial fusion, where two mitochondria fuse together to form a larger one. Fusion can result in elongated mitochondrial networks. In healthy cells, mitochondrial fission and fusion are balanced, and disruptions to these processes are linked to various diseases. Mitochondrial fission is coordinated with the mitochondrial DNA replication process. Some of the proteins involved in mitochondrial fission have been identified, and mutations in some of these proteins are associated with mitochondrial diseases. Mitochondrial fission plays a role in the cellular stress response and in apoptosis.
Eps15 homology domain-containing protein 3, abbreviated as EHD3 and also known as PAST3, is a protein encoded by the EHD3 gene. It has been observed in humans, mice and rats. It belongs to the EHD protein family, a group of four membrane remodeling proteins related to the Dynamin superfamily of large GTPases. Although the four of them are 70-80% amino acid identical, they all have different locations. Its main function is related to endocytic transport.
Dynamin Superfamily Protein (DSP) is a protein superfamily includes classical dynamins, GBPs, Mx proteins, OPA1, mitofusins in Eukaryote, and bacterial dynamin-like proteins (BDLPs) in Prokaryote. DSPs mediate eukaryotic membrane fusion and fission necessary for endocytosis, organelle biogenesis and maintenance, Mitochondrial fusion and fission, as well as for prokaryotic cytokinesis.
Clathrin-independent endocytosis refers to the cellular process by which cells internalize extracellular molecules and particles through mechanisms that do not rely on the protein clathrin, playing a crucial role in diverse physiological processes such as nutrient uptake, membrane turnover, and cellular signaling.