The genome and proteins of HIV (human immunodeficiency virus) have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.

Envelope glycoprotein GP120 is a glycoprotein exposed on the surface of the HIV envelope. It was discovered by Professors Tun-Hou Lee and Myron "Max" Essex of the Harvard School of Public Health in 1988. The 120 in its name comes from its molecular weight of 120 kDa. Gp120 is essential for virus entry into cells as it plays a vital role in attachment to specific cell surface receptors. These receptors are DC-SIGN, Heparan Sulfate Proteoglycan and a specific interaction with the CD4 receptor, particularly on helper T-cells. Binding to CD4 induces the start of a cascade of conformational changes in gp120 and gp41 that lead to the fusion of the viral membrane with the host cell membrane. Binding to CD4 is mainly electrostatic although there are van der Waals interactions and hydrogen bonds.
The murine leukemia viruses are retroviruses named for their ability to cause cancer in murine (mouse) hosts. Some MLVs may infect other vertebrates. MLVs include both exogenous and endogenous viruses. Replicating MLVs have a positive sense, single-stranded RNA (ssRNA) genome that replicates through a DNA intermediate via the process of reverse transcription.
Human foamy virus (HFV) is a retrovirus and specifically belongs to the genus Spumavirus. The spumaviruses are complex and significantly different from the other six genera of retroviruses in several ways. The foamy viruses derive their name from the characteristic ‘foamy’ appearance of the cytopathic effect (CPE) induced in the cells. Foamy virus in humans occurs only as a result of zoonotic infection.

APOBEC3G is a human enzyme encoded by the APOBEC3G gene that belongs to the APOBEC superfamily of proteins. This family of proteins has been suggested to play an important role in innate anti-viral immunity. APOBEC3G belongs to the family of cytidine deaminases that catalyze the deamination of cytidine to uridine in the single stranded DNA substrate. The C-terminal domain of A3G renders catalytic activity, several NMR and crystal structures explain the substrate specificity and catalytic activity.
Simian foamy virus (SFV) is a species of the genus Spumavirus that belongs to the family of Retroviridae. It has been identified in a wide variety of primates, including prosimians, New World and Old World monkeys, as well as apes, and each species has been shown to harbor a unique (species-specific) strain of SFV, including African green monkeys, baboons, macaques, and chimpanzees. As it is related to the more well-known retrovirus human immunodeficiency virus (HIV), its discovery in primates has led to some speculation that HIV may have been spread to the human species in Africa through contact with blood from apes, monkeys, and other primates, most likely through bushmeat-hunting practices.

Endoplasmic-reticulum-associated protein degradation (ERAD) designates a cellular pathway which targets misfolded proteins of the endoplasmic reticulum for ubiquitination and subsequent degradation by a protein-degrading complex, called the proteasome.
Env is a viral gene that encodes the protein forming the viral envelope. The expression of the env gene enables retroviruses to target and attach to specific cell types, and to infiltrate the target cell membrane.
Visna-maedi virus from the genus Lentivirus and subfamily Orthoretrovirinae, is a retrovirus that causes encephalitis and chronic pneumonitis in sheep. It is known as visna when found in the brain, and maedi when infecting the lungs. Lifelong, persistent infections in sheep occur in the lungs, lymph nodes, spleen, joints, central nervous system, and mammary glands; The condition is sometimes known as ovine progressive pneumonia (OPP), particularly in the United States, or Montana sheep disease. White blood cells of the monocyte/macrophage lineage are the main target of the virus.

CD81 molecule, also known as CD81, is a protein which in humans is encoded by the CD81 gene. It is also known as 26 kDa cell surface protein, TAPA-1, and Tetraspanin-28 (Tspan-28).

AP-1 complex subunit gamma-like 2 is a protein that in humans is encoded by the AP1G2 gene.

AP-1 complex subunit sigma-2 is a protein that in humans is encoded by the AP1S2 gene.

Interferon-induced transmembrane protein 1 is a protein that in humans is encoded by the IFITM1 gene. IFITM1 has also recently been designated CD225. This protein has several additional names: fragilis, IFI17 [interferon-induced protein 17], 9-27 [Interferon-inducible protein 9-27] and Leu13.

Tetherin, also known as bone marrow stromal antigen 2, is a lipid raft associated protein that in humans is encoded by the BST2 gene. In addition, tetherin has been designated as CD317. This protein is constitutively expressed in mature B cells, plasma cells and plasmacytoid dendritic cells, and in many other cells, it is only expressed as a response to stimuli from IFN pathway.

Rev is a transactivating protein that is essential to the regulation of HIV-1 protein expression. A nuclear localization signal is encoded in the rev gene, which allows the Rev protein to be localized to the nucleus, where it is involved in the export of unspliced and incompletely spliced mRNAs. In the absence of Rev, mRNAs of the HIV-1 late (structural) genes are retained in the nucleus, preventing their translation.
2F5 is a broadly neutralizing human monoclonal antibody (mAb) that has been shown to bind to and neutralize HIV-1 in vitro, making it a potential candidate for use in vaccine synthesis. 2F5 recognizes an epitope in the membrane-proximal external region (MPER) of HIV-1 gp41. 2F5 then binds to this epitope and its constant region interacts with the viral lipid membrane, which neutralizes the virus.
Vpx is a virion-associated protein encoded by human immunodeficiency virus type 2 HIV-2 and most simian immunodeficiency virus (SIV) strains, but that is absent from HIV-1. It is similar in structure to the protein Vpr that is carried by SIV and HIV-2 as well as HIV-1. Vpx is one of five accessory proteins carried by lentiviruses that enhances viral replication by inhibiting host antiviral factors.
Éric A. Cohen is a Canadian molecular virologist whose research is focused on human immunodeficiency virus (HIV)-host interactions that govern viral replication and persistence.
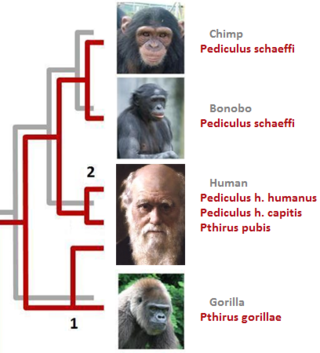
In parasitology and epidemiology, a host switch is an evolutionary change of the host specificity of a parasite or pathogen. For example, the human immunodeficiency virus used to infect and circulate in non-human primates in West-central Africa, but switched to humans in the early 20th century.

Serine incorporator 5 is a protein that in humans is encoded by the SERINC5 gene.











