Interferon regulatory factor 3, also known as IRF3, is an interferon regulatory factor. [5]
Interferon regulatory factor 3, also known as IRF3, is an interferon regulatory factor. [5]
IRF3 is a member of the interferon regulatory transcription factor (IRF) family. [5] IRF3 was originally discovered as a homolog of IRF1 and IRF2. IRF3 has been further characterized and shown to contain several functional domains including a nuclear export signal, a DNA-binding domain, a C-terminal IRF association domain and several regulatory phosphorylation sites. [6] IRF3 is found in an inactive cytoplasmic form that upon serine/threonine phosphorylation forms a complex with CREBBP. [7] The complex translocates into the nucleus for the transcriptional activation of interferons alpha and beta, and further interferon-induced genes. [8]
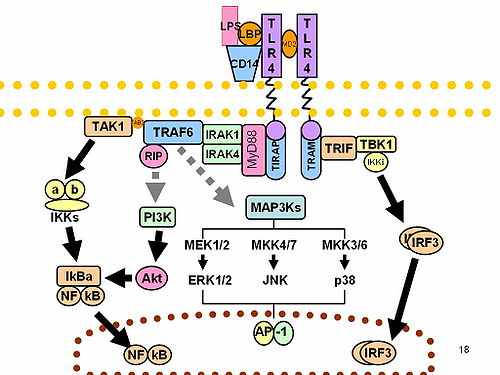
IRF3 plays an important role in the innate immune system's response to viral infection. [9] Aggregated MAVS have been found to activate IRF3 dimerization. [10] A 2015 study shows phosphorylation of innate immune adaptor proteins MAVS, STING and TRIF at a conserved pLxIS motif recruits and specifies IRF3 phosphorylation and activation by the Serine/threonine-protein kinase TBK1, thereby activating the production of type-I interferons. [11] Another study has shown that IRF3-/- knockouts protect from myocardial infarction. [12] The same study identified IRF3 and the type I IFN response as a potential therapeutic target for post-myocardial infarction cardioprotection. [12]

Interferon regulatory factors (IRF) are proteins which regulate transcription of interferons. Interferon regulatory factors contain a conserved N-terminal region of about 120 amino acids, which folds into a structure that binds specifically to the IRF-element (IRF-E) motifs, which is located upstream of the interferon genes. Some viruses have evolved defense mechanisms that regulate and interfere with IRF functions to escape the host immune system. For instance, the remaining parts of the interferon regulatory factor sequence vary depending on the precise function of the protein. The Kaposi sarcoma herpesvirus, KSHV, is a cancer virus that encodes four different IRF-like genes; including vIRF1, which is a transforming oncoprotein that inhibits type 1 interferon activity. In addition, the expression of IRF genes is under epigenetic regulation by promoter DNA methylation.

Signal transducer and activator of transcription 2 is a protein that in humans is encoded by the STAT2 gene. It is a member of the STAT protein family. This protein is critical to the biological response of type I interferons (IFNs). STAT2 sequence identity between mouse and human is only 68%.
NSP1 (NS53), the product of rotavirus gene 5, is a nonstructural RNA-binding protein that contains a cysteine-rich region and is a component of early replication intermediates. RNA-folding predictions suggest that this region of the NSP1 mRNA can interact with itself, producing a stem-loop structure similar to that found near the 5'-terminus of the NSP1 mRNA.

Interferon regulatory factor 2 is a protein that in humans is encoded by the IRF2 gene.

Interferon regulatory factor 7, also known as IRF7, is a member of the interferon regulatory factor family of transcription factors.

RIG-I is a cytosolic pattern recognition receptor (PRR) responsible for the type-1 interferon (IFN1) response. RIG-I is an essential molecule in the innate immune system for recognizing cells that have been infected with a virus. These viruses can include West Nile virus, Japanese Encephalitis virus, influenza A, Sendai virus, flavivirus, and coronaviruses. RIG-I is structurally considered a helical ATP-dependent DExD/H box RNA helicase, that recognizes short viral double-stranded RNA (dsRNA) in the cytosol during a viral infection or other irregular RNAs. Once activated by the dsRNA, the N-terminus caspase activation and recruitment domains (CARDs) migrate and bind with CARDs attached to mitochondrial antiviral signaling protein (MAVS) to activate the signaling pathway for IFN1. IFN1s have three main functions: to limit the virus from spreading to nearby cells, promote an innate immune response, including inflammatory responses, and help activate the adaptive immune system. Other studies have shown that in different microenvironments, such as in cancerous cells, RIG-I has more functions other than viral recognition. RIG-I orthologs are found in mammals, geese, ducks, some fish, and some reptiles. RIG-I is in most cells, including various innate immune system cells, and is usually in an inactive state. Knockout mice that have been designed to have a deleted or non-functioning RIG-I gene are not healthy and typically die embryonically. If they survive, the mice have serious developmental dysfunction. The stimulator of interferon genes STING antagonizes RIG-1 by binding its N-terminus, probably as to avoid overactivation of RIG-1 signaling and the associated autoimmunity.
Interferon regulatory factor 1 is a protein that in humans is encoded by the IRF1 gene.

TBK1 is an enzyme with kinase activity. Specifically, it is a serine / threonine protein kinase. It is encoded by the TBK1 gene in humans. This kinase is mainly known for its role in innate immunity antiviral response. However, TBK1 also regulates cell proliferation, apoptosis, autophagy, and anti-tumor immunity. Insufficient regulation of TBK1 activity leads to autoimmune, neurodegenerative diseases or tumorigenesis.

Interferon alpha-2 is a protein that in humans is encoded by the IFNA2 gene.

Inhibitor of nuclear factor kappa-B kinase subunit epsilon also known as I-kappa-B kinase epsilon or IKK-epsilon is an enzyme that in humans is encoded by the IKBKE gene.

Interferon regulatory factor 4 (IRF4) also known as MUM1 is a protein that in humans is encoded by the IRF4 gene, located at 6p25-p23. IRF4 functions as a key regulatory transcription factor in the development of human immune cells. The expression of IRF4 is essential for the differentiation of T lymphocytes and B lymphocytes as well as certain myeloid cells.

Interferon regulatory factor 9 is a protein that in humans is encoded by the IRF9 gene, previously known as ISGF3G.

Interferon regulatory factor 5 is a protein that in humans is encoded by the IRF5 gene. The IRF family is a group of transcription factors that are involved in signaling for virus responses in mammals along with regulation of certain cellular functions.

Interferon regulatory factor 8 (IRF8) also known as interferon consensus sequence-binding protein (ICSBP), is a protein that in humans is encoded by the IRF8 gene. IRF8 is a transcription factor that plays critical roles in the regulation of lineage commitment and in myeloid cell maturation including the decision for a common myeloid progenitor (CMP) to differentiate into a monocyte precursor cell.

Mitochondrial antiviral-signaling protein (MAVS) is a protein that is essential for antiviral innate immunity. MAVS is located in the outer membrane of the mitochondria, peroxisomes, and mitochondrial-associated endoplasmic reticulum membrane (MAM). Upon viral infection, a group of cytosolic proteins will detect the presence of the virus and bind to MAVS, thereby activating MAVS. The activation of MAVS leads the virally infected cell to secrete cytokines. This induces an immune response which kills the host's virally infected cells, resulting in clearance of the virus.
RIG-like receptors are a type of intracellular pattern recognition receptor involved in the recognition of viruses by the innate immune system. RIG-I is the best characterized receptor within the RIG-I like receptor (RLR) family. Together with MDA5 and LGP2, this family of cytoplasmic pattern recognition receptors (PRRs) are sentinels for intracellular viral RNA that is a product of viral infection. The RLR receptors provide frontline defence against viral infections in most tissues.

Phosphomimetics are amino acid substitutions that mimic a phosphorylated protein, thereby activating the protein. Within cells, proteins are commonly modified at serine, tyrosine and threonine amino acids by adding a phosphate group. Phosphorylation is a common mode of activating or deactivating a protein as a form of regulation. However some non-phosphorylated amino acids appear chemically similar to phosphorylated amino acids. Therefore, by replacing an amino acid, the protein may maintain a higher level of activity. For example, aspartic acid is chemically similar to phospho-serine. Therefore, when an aspartic acid replaces a serine, it is a phosphomimetic of phospho-serine and can make the protein always in its phosphorylated form. Phosphonate-based compounds have been used as phosphotyrosine analogues, as they are less enzyme labile and are physiologically more stable.
The interleukin-1 receptor (IL-1R) associated kinase (IRAK) family plays a crucial role in the protective response to pathogens introduced into the human body by inducing acute inflammation followed by additional adaptive immune responses. IRAKs are essential components of the Interleukin-1 receptor signaling pathway and some Toll-like receptor signaling pathways. Toll-like receptors (TLRs) detect microorganisms by recognizing specific pathogen-associated molecular patterns (PAMPs) and IL-1R family members respond the interleukin-1 (IL-1) family cytokines. These receptors initiate an intracellular signaling cascade through adaptor proteins, primarily, MyD88. This is followed by the activation of IRAKs. TLRs and IL-1R members have a highly conserved amino acid sequence in their cytoplasmic domain called the Toll/Interleukin-1 (TIR) domain. The elicitation of different TLRs/IL-1Rs results in similar signaling cascades due to their homologous TIR motif leading to the activation of mitogen-activated protein kinases (MAPKs) and the IκB kinase (IKK) complex, which initiates a nuclear factor-κB (NF-κB) and AP-1-dependent transcriptional response of pro-inflammatory genes. Understanding the key players and their roles in the TLR/IL-1R pathway is important because the presence of mutations causing the abnormal regulation of Toll/IL-1R signaling leading to a variety of acute inflammatory and autoimmune diseases.
The cGAS–STING pathway is a component of the innate immune system that functions to detect the presence of cytosolic DNA and, in response, trigger expression of inflammatory genes that can lead to senescence or to the activation of defense mechanisms. DNA is normally found in the nucleus of the cell. Localization of DNA to the cytosol is associated with tumorigenesis, viral infection, and invasion by some intracellular bacteria. The cGAS – STING pathway acts to detect cytosolic DNA and induce an immune response.

Interferon regulatory factor 2 binding protein 2 is a protein that in humans is encoded by the IRF2BP2 gene.