Related Research Articles

The enzyme fructose bisphosphatase (EC 3.1.3.11; systematic name D-fructose-1,6-bisphosphate 1-phosphohydrolase) catalyses the conversion of fructose-1,6-bisphosphate to fructose 6-phosphate in gluconeogenesis and the Calvin cycle, which are both anabolic pathways:
Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the biomolecule of interest; antigen and antibody, enzyme and substrate, receptor and ligand, or protein and nucleic acid binding interactions are frequently exploited for isolation of various biomolecules. Affinity chromatography is useful for its high selectivity and resolution of separation, compared to other chromatographic methods.

Oxaloacetate decarboxylase is a carboxy-lyase involved in the conversion of oxaloacetate into pyruvate.

α-Glucosidase is a glucosidase located in the brush border of the small intestine that acts upon α(1→4) bonds:

Protein C inhibitor is a serine protease inhibitor (serpin) that limits the activity of protein C.
In enzymology, a tryptophan alpha,beta-oxidase (EC 1.3.3.10) is an enzyme that catalyzes the chemical reaction
In enzymology, a mucinaminylserine mucinaminidase (EC 3.2.1.110) is an enzyme that catalyzes the chemical reaction
In enzymology, a beta-galactoside alpha-2,6-sialyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a sucrose-1,6-alpha-glucan 3(6)-alpha-glucosyltransferase is an enzyme that catalyzes the chemical reaction
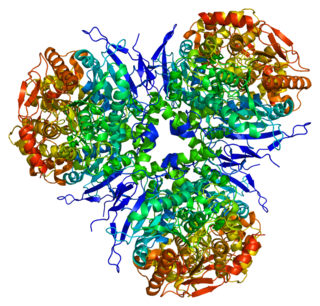
Liver carboxylesterase 1 also known as carboxylesterase 1 is an enzyme that in humans is encoded by the CES1 gene. The protein is also historically known as serine esterase 1 (SES1), monocyte esterase and cholesterol ester hydrolase (CEH). Three transcript variants encoding three different isoforms have been found for this gene. The various protein products from isoform a, b and c range in size from 568, 567 and 566 amino acids long, respectively.

Pterin-4-alpha-carbinolamine dehydratase is an enzyme that in humans is encoded by the PCBD1 gene.

3α-Hydroxysteroid dehydrogenase is an enzyme that in humans is encoded by the AKR1C4 gene. It is known to be necessary for the synthesis of the endogenous neurosteroids allopregnanolone, THDOC, and 3α-androstanediol. It is also known to catalyze the reversible conversion of 3α-androstanediol (5α-androstane-3α,17β-diol) to dihydrotestosterone and vice versa.

N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase is an enzyme that in humans is encoded by the NAGPA gene.
Agarase is an enzyme with systematic name agarose 4-glycanohydrolase. It is found in agarolytic bacteria and is the first enzyme in the agar catabolic pathway. It is responsible for allowing them to use agar as their primary source of carbon and enables their ability to thrive in the ocean.

Flavocytochrome c sulfide dehydrogenase, also known as Sulfide-cytochrome-c reductase (flavocytochrome c) (EC 1.8.2.3), is an enzyme with systematic name hydrogen-sulfide:flavocytochrome c oxidoreductase. It is found in sulfur-oxidising bacteria such as the purple phototrophic bacteria Allochromatium vinosum. This enzyme catalyses the following chemical reaction:
5β-Reductase, or Δ4-3-oxosteroid 5β-reductase (EC 1.3.1.3, 3-oxo-Δ4-steroid 5β-reductase, androstenedione 5β-reductase, cholestenone 5β-reductase, cortisone 5β-reductase, cortisone Δ4-5β-reductase, steroid 5β-reductase, testosterone 5β-reductase, Δ4-3-ketosteroid 5β-reductase, Δ4-5β-reductase, Δ4-hydrogenase, 4,5β-dihydrocortisone:NADP+ Δ4-oxidoreductase, 3-oxo-5β-steroid:NADP+ Δ4-oxidoreductase) is an enzyme with systematic name 5β-cholestan-3-one:NADP+ 4,5-oxidoreductase. This enzyme catalyses the following chemical reaction
(N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase is an enzyme with systematic name UDP-N-acetyl-D-galactosamine:1-O-(O- - -O-beta-D-galactopyranosyl- -beta-D-glucopyranosyl)-ceramide 4-beta-N-acetyl-D-galactosaminyltransferase. This enzyme catalyses the following chemical reaction
Phospholipase C (EC 3.1.4.3, lipophosphodiesterase I, Clostridium welchii α-toxin, Clostridium oedematiens β- and γ-toxins, lipophosphodiesterase C, phosphatidase C, heat-labile hemolysin, α-toxin) is an enzyme with systematic name phosphatidylcholine cholinephosphohydrolase. This enzyme catalyses the following chemical reaction
4-alpha-D-{(1->4)-alpha-D-glucano}trehalose trehalohydrolase is an enzyme with systematic name 4-alpha-D-( -alpha-D-glucano)trehalose glucanohydrolase (trehalose-producing). This enzyme catalyses the following chemical reaction
The enzyme exo-(1→4)-α-D-glucan lyase (EC 4.2.2.13, α-(1→4)-glucan 1,5-anhydro-D-fructose eliminase, α-1,4-glucan exo-lyase, α-1,4-glucan lyase, GLase) is an enzyme with systematic name (1→4)-α-D-glucan exo-4-lyase (1,5-anhydro-D-fructose-forming). This enzyme catalyses the following chemical reaction
References
- ↑ Potin P, Richard C, Rochas C, Kloareg B (June 1993). "Purification and characterization of the alpha-agarase from Alteromonas agarlyticus (Cataldi) comb. nov., strain GJ1B". European Journal of Biochemistry. 214 (2): 599–607. doi: 10.1111/j.1432-1033.1993.tb17959.x . PMID 8513809.
- ↑ Ohta Y, Hatada Y, Miyazaki M, Nogi Y, Ito S, Horikoshi K (April 2005). "Purification and characterization of a novel alpha-agarase from a Thalassomonas sp". Current Microbiology. 50 (4): 212–6. doi:10.1007/s00284-004-4435-z. PMID 15902469.