4-Aminobenzoic acid (also known as para-aminobenzoic acid or PABA because the two functional groups are attached to the benzene ring across from one another in the para position) is an organic compound with the formula H2NC6H4CO2H. PABA is a white solid, although commercial samples can appear gray. It is slightly soluble in water. It consists of a benzene ring substituted with amino and carboxyl groups. The compound occurs extensively in the natural world.
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.
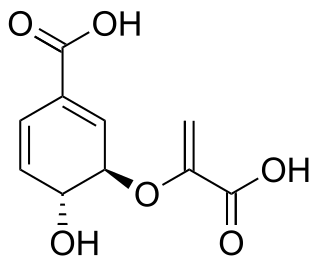
Chorismic acid, more commonly known as its anionic form chorismate, is an important biochemical intermediate in plants and microorganisms. It is a precursor for:

Amino acid synthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids).
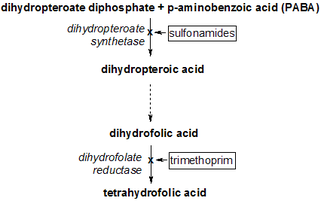
Dihydropteroate synthase is an enzyme classified under EC 2.5.1.15. It produces dihydropteroate in bacteria, but it is not expressed in most eukaryotes including humans. This makes it a useful target for sulfonamide antibiotics, which compete with the PABA precursor.
CTP synthase is an enzyme involved in pyrimidine biosynthesis that interconverts UTP and CTP.

Isochorismate synthase ( EC 5.4.4.2) is an isomerase enzyme that catalyzes the first step in the biosynthesis of vitamin K2 (menaquinone) in Escherichia coli.

In enzymology, a phosphoribosylanthranilate isomerase (PRAI) is an enzyme that catalyzes the third step of the synthesis of the amino acid tryptophan.

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction
4-amino-4-deoxychorismate lyase is an enzyme that participates in folate biosynthesis by catalyzing the production of PABA by the following reaction

The enzyme anthranilate synthase catalyzes the chemical reaction

The enzyme chorismate lyase catalyzes the first step in ubiquinone biosynthesis, the removal of pyruvate from chorismate, to yield 4-hydroxybenzoate in Escherichia coli and other Gram-negative bacteria. It belongs to the family of lyases, specifically the oxo-acid-lyases, which cleave carbon-carbon bonds. The systematic name of this enzyme class is chorismate pyruvate-lyase (4-hydroxybenzoate-forming). Other names in common use include CL, CPL, and UbiC.
The enzyme indole-3-glycerol-phosphate synthase (IGPS) (EC 4.1.1.48) catalyzes the chemical reaction
The enzyme chorismate synthase catalyzes the chemical reaction

In enzymology, a malate synthase (EC 2.3.3.9) is an enzyme that catalyzes the chemical reaction

In enzymology, a cystathionine gamma-synthase is an enzyme that catalyzes the formation of cystathionine from cysteine and an activated derivative of homoserine, e.g.:

In enzymology, a 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase is an enzyme that catalyzes the chemical reaction

In molecular biology, glutamine amidotransferases (GATase) are enzymes which catalyse the removal of the ammonia group from a glutamine molecule and its subsequent transfer to a specific substrate, thus creating a new carbon-nitrogen group on the substrate. This activity is found in a range of biosynthetic enzymes, including glutamine amidotransferase, anthranilate synthase component II, p-aminobenzoate, and glutamine-dependent carbamoyl-transferase (CPSase). Glutamine amidotransferase (GATase) domains can occur either as single polypeptides, as in glutamine amidotransferases, or as domains in a much larger multifunctional synthase protein, such as CPSase. On the basis of sequence similarities two classes of GATase domains have been identified: class-I and class-II. Class-I GATase domains are defined by a conserved catalytic triad consisting of cysteine, histidine and glutamate. Class-I GATase domains have been found in the following enzymes: the second component of anthranilate synthase and 4-amino-4-deoxychorismate (ADC) synthase; CTP synthase; GMP synthase; glutamine-dependent carbamoyl-phosphate synthase; phosphoribosylformylglycinamidine synthase II; and the histidine amidotransferase hisH.

3-Deoxy-D-arabino-heptulosonic acid 7-phosphate (DAHP) is a 7-carbon ulonic acid. This compound is found in the shikimic acid biosynthesis pathway and is an intermediate in the production of aromatic amino acids.
2-amino-4-deoxychorismate synthase is an enzyme with systematic name (2S)-2-amino-4-deoxychorismate:2-oxoglutarate aminotransferase. This enzyme catalyses the following chemical reaction


















