
The citric acid cycle —also known as the Krebs cycle, Szent-Györgyi-Krebs cycle or the TCA cycle (tricarboxylic acid cycle)—is a series of chemical reactions to release stored energy through the oxidation of acetyl-CoA derived from carbohydrates, fats, and proteins. The Krebs cycle is used by organisms that respire (as opposed to organisms that ferment) to generate energy, either by anaerobic respiration or aerobic respiration. In addition, the cycle provides precursors of certain amino acids, as well as the reducing agent NADH, that are used in numerous other reactions. Its central importance to many biochemical pathways suggests that it was one of the earliest components of metabolism. Even though it is branded as a 'cycle', it is not necessary for metabolites to follow only one specific route; at least three alternative segments of the citric acid cycle have been recognized.
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.

Succinyl coenzyme A synthetase is an enzyme that catalyzes the reversible reaction of succinyl-CoA to succinate. The enzyme facilitates the coupling of this reaction to the formation of a nucleoside triphosphate molecule from an inorganic phosphate molecule and a nucleoside diphosphate molecule. It plays a key role as one of the catalysts involved in the citric acid cycle, a central pathway in cellular metabolism, and it is located within the mitochondrial matrix of a cell.
Glutathione synthetase (GSS) is the second enzyme in the glutathione (GSH) biosynthesis pathway. It catalyses the condensation of gamma-glutamylcysteine and glycine, to form glutathione. Glutathione synthetase is also a potent antioxidant. It is found in many species including bacteria, yeast, mammals, and plants.

In enzymology, a dihydrofolate synthase is an enzyme that catalyzes the chemical reaction
Glutamate–cysteine ligase (GCL) EC 6.3.2.2), previously known as γ-glutamylcysteine synthetase (GCS), is the first enzyme of the cellular glutathione (GSH) biosynthetic pathway that catalyzes the chemical reaction:

In molecular biology, the protein domain SAICAR synthase is an enzyme which catalyses a reaction to create SAICAR. In enzymology, this enzyme is also known as phosphoribosylaminoimidazolesuccinocarboxamide synthase. It is an enzyme that catalyzes the chemical reaction

In enzymology, a succinate—CoA ligase (GDP-forming) is an enzyme that catalyzes the chemical reaction
In enzymology, a tetrahydrofolate synthase is an enzyme that catalyzes the chemical reaction
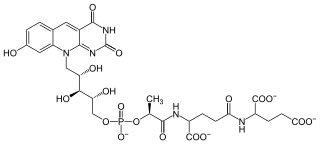
Coenzyme F420 or 8-hydroxy-5-deazaflavin is a coenzyme (sometimes called a cofactor) involved in redox reactions in methanogens, in many Actinomycetota, and sporadically in other bacterial lineages. It is a flavin derivative with an absorption maximum at 420 nm—hence its name. The coenzyme is a substrate for coenzyme F420 hydrogenase, 5,10-methylenetetrahydromethanopterin reductase and methylenetetrahydromethanopterin dehydrogenase.

L-2-hydroxycarboxylate dehydrogenase (NAD+) (EC 1.1.1.337, (R)-sulfolactate:NAD+ oxidoreductase, L-sulfolactate dehydrogenase, (R)-sulfolactate dehydrogenase, L-2-hydroxyacid dehydrogenase (NAD+), ComC) is an enzyme with systematic name (2S)-2-hydroxycarboxylate:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
Glucose-6-phosphate dehydrogenase (coenzyme-F420) is an enzyme with systematic name D-glucose-6-phosphate:F420 1-oxidoreductase. This enzyme catalyses the following chemical reaction
7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase (EC 4.3.1.32, FO synthase) and 5-amino-6-(D-ribitylamino)uracil—L-tyrosine 4-hydroxyphenyl transferase (EC 2.5.1.147) are two enzymes always complexed together to achieve synthesis of FO, a precursor to Coenzyme F420. Their systematic names are 5-amino-5-(4-hydroxybenzyl)-6-(D-ribitylimino)-5,6-dihydrouracil ammonia-lyase (7,8-didemethyl-8-hydroxy-5-deazariboflavin-forming) and 5-amino-6-(D-ribitylamino)uracil:L-tyrosine, 4-hydroxyphenyl transferase respectively. The enzymes catalyse the following chemical reactions:
2-Phospho-L-lactate guanylyltransferase is an enzyme with systematic name GTP:2-phospho-L-lactate guanylyltransferase. This enzyme catalyses the following chemical reaction
2-phospho-L-lactate transferase is an enzyme with systematic name (2S)-lactyl-2-diphospho-5'-guanosine:7,8-didemethyl-8-hydroxy-5-deazariboflavin 2-phospho-L-lactate transferase. This enzyme catalyses the following chemical reaction
Coenzyme gamma-F420-2:α-L-glutamate ligase (EC 6.3.2.32, MJ1001, CofF protein, gamma-F420-2:alpha-L-glutamate ligase) is an enzyme with systematic name L-glutamate:coenzyme gamma-F420-2 (ADP-forming). This enzyme catalyses the following chemical reaction
Tetrahydrosarcinapterin synthase is an enzyme with systematic name tetrahydromethanopterin:alpha-L-glutamate ligase (ADP-forming). This enzyme catalyses the following chemical reaction
Coenzyme F420-1:γ-L-glutamate ligase (EC 6.3.2.34, F420:gamma-glutamyl ligase, CofE-AF, MJ0768, CofE) is an enzyme with systematic name L-glutamate:coenzyme F420-1 ligase (GDP-forming). This enzyme catalyses the following chemical reaction







