
Pyruvate dehydrogenase complex (PDC) is a complex of three enzymes that converts pyruvate into acetyl-CoA by a process called pyruvate decarboxylation. Acetyl-CoA may then be used in the citric acid cycle to carry out cellular respiration, and this complex links the glycolysis metabolic pathway to the citric acid cycle. Pyruvate decarboxylation is also known as the "pyruvate dehydrogenase reaction" because it also involves the oxidation of pyruvate.
The branched-chain α-ketoacid dehydrogenase complex is a multi-subunit complex of enzymes that is found on the mitochondrial inner membrane. This enzyme complex catalyzes the oxidative decarboxylation of branched, short-chain alpha-ketoacids. BCKDC is a member of the mitochondrial α-ketoacid dehydrogenase complex family, which also includes pyruvate dehydrogenase and alpha-ketoglutarate dehydrogenase, key enzymes that function in the Krebs cycle.

Dihydrolipoyl transacetylase is an enzyme component of the multienzyme pyruvate dehydrogenase complex. The pyruvate dehydrogenase complex is responsible for the pyruvate decarboxylation step that links glycolysis to the citric acid cycle. This involves the transformation of pyruvate from glycolysis into acetyl-CoA which is then used in the citric acid cycle to carry out cellular respiration.
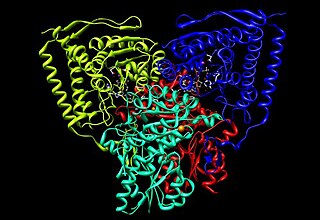
Pyruvate dehydrogenase is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. The conversion requires the coenzyme thiamine pyrophosphate.

In enzymology, a malate dehydrogenase (NADP+) (EC 1.1.1.82) is an enzyme that catalyzes the chemical reaction

In enzymology, a 3-methyl-2-oxobutanoate dehydrogenase (EC 1.2.4.4) is an enzyme that catalyzes the chemical reaction
In enzymology, a saccharopine dehydrogenase (NAD+, L-glutamate-forming) (EC 1.5.1.9) is an enzyme that catalyzes the chemical reaction
In enzymology, a saccharopine dehydrogenase (NADP+, L-lysine-forming) (EC 1.5.1.8) is an enzyme that catalyzes the chemical reaction
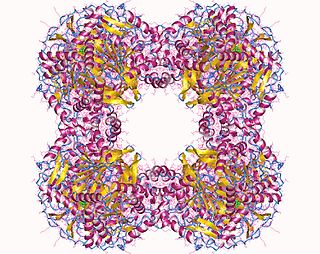
In enzymology, a dihydrolipoyllysine-residue succinyltransferase (EC 2.3.1.61) is an enzyme that catalyzes the chemical reaction
In enzymology, a lipoyl(octanoyl) transferase (EC 2.3.1.181) is an enzyme that catalyzes the chemical reaction
In enzymology, a [3-methyl-2-oxobutanoate dehydrogenase (acetyl-transferring)] is an enzyme that catalyzes the chemical reaction

A 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial is an enzyme that in humans is encoded by the BCKDHA gene.

Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial is an enzyme that in humans is encoded by the DBT gene.

Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial is an enzyme that in humans is encoded by the DLST gene.

2-Oxoisovalerate dehydrogenase subunit beta, mitochondrial is an enzyme that in humans is encoded by the BCKDHB gene.

Branched chain ketoacid dehydrogenase kinase (BCKDK) is an enzyme encoded by the BCKDK gene on chromosome 16. This enzyme is part of the mitochondrial protein kinases family and it is a regulator of the valine, leucine, and isoleucine catabolic pathways. BCKDK is found in the mitochondrial matrix and the prevalence of it depends on the type of cell. Liver cells tend to have the lowest concentration of BCKDK, whereas skeletal muscle cells have the highest amount. Abnormal activity of this enzyme often leads to diseases such as maple syrup urine disease and cachexia.

Alpha-aminoadipic semialdehyde synthase is an enzyme encoded by the AASS gene in humans and is involved in their major lysine degradation pathway. It is similar to the separate enzymes coded for by the LYS1 and LYS9 genes in yeast, and related to, although not similar in structure, the bifunctional enzyme found in plants. In humans, mutations in the AASS gene, and the corresponding alpha-aminoadipic semialdehyde synthase enzyme are associated with familial hyperlysinemia. This rare disease is inherited in an autosomal recessive pattern and patients often have no clinical symptoms.
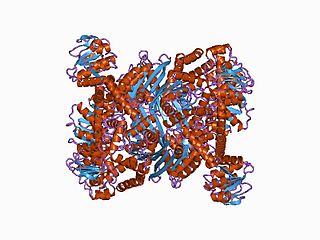
In molecular biology, the ELFV dehydrogenase family of enzymes include glutamate, leucine, phenylalanine and valine dehydrogenases. These enzymes are structurally and functionally related. They contain a Gly-rich region containing a conserved Lys residue, which has been implicated in the catalytic activity, in each case a reversible oxidative deamination reaction.
Lipoate–protein ligase (EC 2.7.7.63, LplA, lipoate protein ligase, lipoate–protein ligase A, LPL, LPL-B) is an enzyme with systematic name ATP:lipoate adenylyltransferase. This enzyme catalyses the following chemical reaction
(3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring))-phosphatase (EC 3.1.3.52, branched-chain oxo-acid dehydrogenase phosphatase, branched-chain 2-keto acid dehydrogenase phosphatase, branched-chain α-keto acid dehydrogenase phosphatase, BCKDH', [3-methyl-2-oxobutanoate dehydrogenase (lipoamide)]-phosphatase, [3-methyl-2-oxobutanoate dehydrogenase (lipoamide)]-phosphate phosphohydrolase) is an enzyme with systematic name (3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring))-phosphate phosphohydrolase. This enzyme catalyses the following chemical reaction














