
Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.
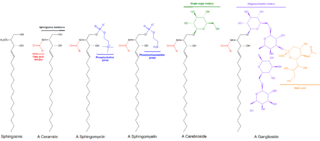
Sphingolipids are a class of lipids containing a backbone of sphingoid bases, which are a set of aliphatic amino alcohols that includes sphingosine. They were discovered in brain extracts in the 1870s and were named after the mythological sphinx because of their enigmatic nature. These compounds play important roles in signal transduction and cell recognition. Sphingolipidoses, or disorders of sphingolipid metabolism, have particular impact on neural tissue. A sphingolipid with a terminal hydroxyl group is a ceramide. Other common groups bonded to the terminal oxygen atom include phosphocholine, yielding a sphingomyelin, and various sugar monomers or dimers, yielding cerebrosides and globosides, respectively. Cerebrosides and globosides are collectively known as glycosphingolipids.
Palmitoyl-CoA is an acyl-CoA thioester. It is an "activated" form of palmitic acid and can be transported into the mitochondrial matrix by the carnitine shuttle system, and once inside can participate in beta-oxidation. Alternatively, palmitoyl-CoA is used as a substrate in the biosynthesis of sphingosine.

Carnitine palmitoyltransferase I (CPT1) also known as carnitine acyltransferase I, CPTI, CAT1, CoA:carnitine acyl transferase (CCAT), or palmitoylCoA transferase I, is a mitochondrial enzyme responsible for the formation of acyl carnitines by catalyzing the transfer of the acyl group of a long-chain fatty acyl-CoA from coenzyme A to l-carnitine. The product is often palmitoylcarnitine, but other fatty acids may also be substrates. It is part of a family of enzymes called carnitine acyltransferases. This "preparation" allows for subsequent movement of the acyl carnitine from the cytosol into the intermembrane space of mitochondria.
Pantothenate kinase (EC 2.7.1.33, PanK; CoaA) is the first enzyme in the Coenzyme A (CoA) biosynthetic pathway. It phosphorylates pantothenate (vitamin B5) to form 4'-phosphopantothenate at the expense of a molecule of adenosine triphosphate (ATP). It is the rate-limiting step in the biosynthesis of CoA.
Palmitoyl-CoA hydrolase (EC 3.1.2.2) is an enzyme in the family of hydrolases that specifically acts on thioester bonds. It catalyzes the hydrolysis of long chain fatty acyl thioesters of acyl carrier protein or coenzyme A to form free fatty acid and the corresponding thiol:

Carnitine O-octanoyltransferase is a member of the transferase family, more specifically a carnitine acyltransferase, a type of enzyme which catalyzes the transfer of acyl groups from acyl-CoAs to carnitine, generating CoA and an acyl-carnitine. Specifically, CROT catalyzes the chemical reaction:

Serine palmitoyltransferase, long chain base subunit 1, also known as SPTLC1, is a protein which in humans is encoded by the SPTLC1 gene.

Phosphoglycerate dehydrogenase (PHGDH) is an enzyme that catalyzes the chemical reactions

Serine palmitoyltransferase, long chain base subunit 2, also known as SPTLC2, is a protein which in humans is encoded by the SPTLC2 gene. SPTLC2 belongs to the class-II pyridoxal-phosphate-dependent aminotransferase family.

Acyl-CoA thioesterase 2, also known as ACOT2, is an enzyme which in humans is encoded by the ACOT2 gene.

Acyl-coenzyme A thioesterase 4 is an enzyme that in humans is encoded by the ACOT4 gene.
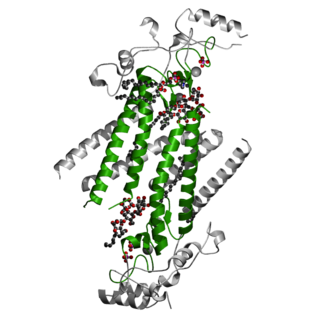
In molecular biology the DHHC domain is a protein domain that acts as an enzyme, which adds a palmitoyl chemical group to proteins in order to anchor them to cell membranes. The DHHC domain was discovered in 1999 and named after a conserved sequence motif found in its protein sequence. Roth and colleagues showed that the yeast Akr1p protein could palmitoylate Yck2p in vitro and inferred that the DHHC domain defined a large family of palmitoyltransferases. In mammals twenty three members of this family have been identified and their substrate specificities investigated. Some members of the family such as ZDHHC3 and ZDHHC7 enhance palmitoylation of proteins such as PSD-95, SNAP-25, GAP43, Gαs. Others such as ZDHHC9 showed specificity only toward the H-Ras protein. However, a recent study questions the involvement of classical enzyme-substrate recognition and specificity in the palmitoylation reaction. Several members of the family have been implicated in human diseases.

Acyl-CoA thioesterase 6 is a protein that in humans is encoded by the ACOT6 gene. The protein, also known as C14orf42, is an enzyme with thioesterase activity.
Very-long-chain 3-oxoacyl-CoA synthase (EC 2.3.1.199, very-long-chain 3-ketoacyl-CoA synthase, very-long-chain beta-ketoacyl-CoA synthase, condensing enzyme, CUT1, CER6, FAE1, KCS, ELO) is an enzyme with systematic name malonyl-CoA:very-long-chain acyl-CoA malonyltransferase (decarboxylating and thioester-hydrolysing). This enzyme catalyses the following chemical reaction
3-hydroxydecanoyl-(acyl-carrier-protein) dehydratase (EC 4.2.1.60, D-3-hydroxydecanoyl-[acyl-carrier protein] dehydratase, 3-hydroxydecanoyl-acyl carrier protein dehydrase, 3-hydroxydecanoyl-acyl carrier protein dehydratase, β-hydroxydecanoyl thioester dehydrase, β-hydroxydecanoate dehydrase, beta-hydroxydecanoyl thiol ester dehydrase, FabA, β-hydroxyacyl-acyl carrier protein dehydratase, HDDase, β-hydroxyacyl-ACP dehydrase, (3R)-3-hydroxydecanoyl-[acyl-carrier-protein] hydro-lyase) is an enzyme with systematic name (3R)-3-hydroxydecanoyl-(acyl-carrier protein) hydro-lyase. This enzyme catalyses the following chemical reaction
Hereditary sensory and autonomic neuropathy type I or hereditary sensory neuropathy type I is a group of autosomal dominant inherited neurological diseases that affect the peripheral nervous system particularly on the sensory and autonomic functions. The hallmark of the disease is the marked loss of pain and temperature sensation in the distal parts of the lower limbs. The autonomic disturbances, if present, manifest as sweating abnormalities.

Acyl-CoA thioesterase 13 is a protein that in humans is encoded by the ACOT13 gene. This gene encodes a member of the thioesterase superfamily. In humans, the protein co-localizes with microtubules and is essential for sustained cell proliferation.
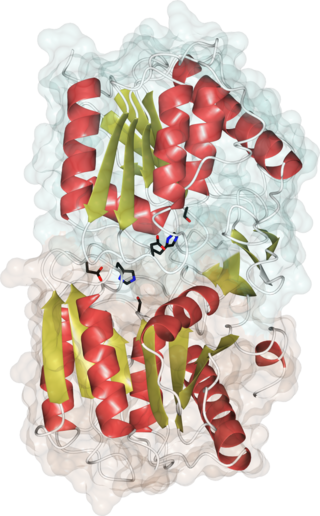
Acyl-protein thioesterases are enzymes that cleave off lipid modifications on proteins, located on the sulfur atom of cysteine residues linked via a thioester bond. Acyl-protein thioesterases are part of the α/β hydrolase superfamily of proteins and have a conserved catalytic triad. For that reason, acyl-protein thioesterases are also able to hydrolyze oxygen-linked ester bonds.
The 1-deoxysphingolipids (1-deoxySLs) are an atypical and recently discovered class of sphingolipids (SLs). They are formed during the nove synthesis pathway and their essential C1-OH deficit causes the malfunctions of the following transformations to achieve complex sphingolipids. In general, sphingolipids are formed during a reaction that is catalyzed by the SPT enzyme (serine-palmitoyltransferase) where the condensation of serine and palmitoyl-CoA takes place. The origin of this rare sphingolipid, though, is due to a defect of the SPT which can also use alanine or glycine. This change is what forms the 1-deoxySL.












