| Metabolite | Major Chemical Shift (ppm) | Function | in vivo MRS Applications | Clinical Applications |
|---|
| N-Acetyl Aspartate (NAA) [19] : 52–53 | 2.01 | - Osmoregulation
- Precursor to neurotransmitter NAAG
- Fatty acid and myelin synthesis (via storage form for acetyl groups)
| Marker of neuronal density Concentration marker | - Believed to reflect neuronal dysfunction rather than neuronal loss
- Elevated levels seen in Canavan's syndrome and sickle cell disease in newborn infants and young children
- Reduced levels seen in chronic stages of stroke, brain tumors, and multiple sclerosis
|
| N-Acetyl Aspartyl Glutamate (NAAG) [19] : 53–54 | 2.04 | - Neurotransmitter involved in excitatory neurotransmission
- Source of glutamate
| Sum of NAA and NAAG provides a reliable estimate of NAA-containing molecules | |
| Adenosine Triphosphate (ATP) [19] : 54–55 | 4.20 - 4.80, 6.13, 8.22 | - Principal donor of free energy in biological systems
| Normally detected with 31P NMR spectroscopy, more difficult to detect by 1H NMR spectroscopy
| - Measure cerebral mitochondrial function
|
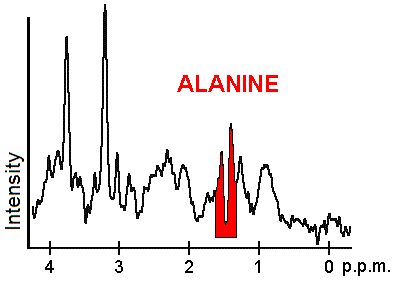
| Alanine (Ala) [19] : 55–56 | 1.40 | - Linked to metabolic pathways like glycolysis, gluconeogensis, and TCA cycle
| None | - Elevated levels seen in melingomas
|
| γ-aminobutyric acid (GABA) [19] : 56–57 | 3.00 | - Inhibitory neurotransmitter
- Regulation of muscle tone
| None | - Reduced levels seen in recent seizures, schizophrenia, autism, bipolar disorder, major depressive disorder
|
| Ascorbic Acid (Asc - Vitamin C) [19] : 57–58 | 4.49 | - Antioxidant
- Coenzyme for the formation of collagen
| Target for hyperpolarized 13C applications to image the redox status in vivo | - Measure ascorbic acid uptake
|
| Aspartic Acid (Asc) [19] : 58 | 3.89 | - Excitatory neurotransmitter
- Metabolite in urea cycle
- Participates in gluconeogenesis
- Essential to malate-aspartate shuttle
| None | |
| Carnitine [19] : 82 | 3.21 | - Transporting long-chain fatty acids across the mitochondrial membrane for β-oxidation
- Regulatory role in substrate switching and glucose homeostasis
| None | - Direct correlation between carnitine content in seminal fluid and sperm count and mobility
- Manage cardiac ischemia and peripheral arterial disease
|
| Carnosine [19] : 84 | 7.09 | - Antioxidant
- Increase Hayflick limit in fibroblasts
- Reduce telomere shortening rate
- Important intramuscular buffer
| Noninvasive method to measure intracellular pH with 1H NMR in vivo | - Reduces development of atherosclerotic buildup
|
| Choline-containing Compounds (tCho) [19] : 59–61 | 3.20 | - Involved in pathways of phospholipid synthesis and degradation
| None | - Elevated levels seen in cancer, Alzheimer's disease, and multiple sclerosis
- Reduced levels are associated with liver disease and stroke
|
| Citric Acid | 2.57, 2.72 | - Intermediate of the Krebs cycle
| None | - Elevated levels in brain tumors
- Diagnosis of malignant adenocarcinoma and benign prostatic hyperplasia
|
| Creatine (Cr) and Phosphocreatine (PCr) [19] : 61–82 | 3.03 | - Energy buffer, retaining constant ATP levels through the creatine kinase reaction
- Energy shuttle, diffusing from energy producing (i.e. mitochondria) to energy utilizing sites (i.e. myofibrils in muscle or nerve terminals in brain)
| None | - Reduced levels seen in chronic phases of many pathologies, including tumors and strokes
|
| Deoxymyoglobin (DMb) [19] : 87 | 79.00 | - Oxygen storage compound and facilitator of oxygen diffusion
| None | - Elevated levels in ischemic conditions (i.e. heavy exercise, using a pressure cuff)
- Determine oxygen saturation in human skeletal and cardiac muscle
|
| Glucose (Glc) [19] : 63 | 5.22 | - Ubiquitous source of energy from bacteria to humans
- Broken down in TCA cycle to provide energy in form of ATP
| Common target in 13C applications to study metabolic pathways | - Elevated levels in people with Alzheimer's
|
| Glutamate (Glu) [19] : 64–65 | 2.20 - 2.40 | - Major excitatory neurotransmitter
- Direct precursor for the major inhibitory neurotransmitter GABA
- Important precursor to synthesis of glutathione
- Glutamate-Glutamine neurotransmitter cycle
| Separation between glutamate and glutamine becomes unreliable, although the sum (Glx) can be quantified with high accuracy | - Elevated levels in bipolar disorder
- Reduced levels in major depressive disorder
|
| Glutamine (Gln) [19] : 65–66 | 2.20 - 2.40 | - Ammonia detoxification
- Glutamate-Glutamine neurotransmitter cycle
| Separation between glutamate and glutamine becomes unreliable, although the sum (Glx) can be quantified with high accuracy | - Elevated levels during hyperammonemia
- Good indicator of liver disease
- Fuel source for a number of cancers
|
| Glutathione (GSH) [19] : 66–67 > | 3.77 | - Antioxidant
- Essential for maintaining normal red-blood-cell structure and keeping hemoglobin in ferrous state
- Storage form of cysteine
| None | - Measure of cellular oxidative stress
- Altered levels in Parkinson's disease and other neurodegenerative diseases affecting the basal ganglia
|
| Glycerol [19] : 67–68 | 3.55, 3.64, 3.77 | - Major constituent of phospholipids
| Difficult to observe in 1H NMR spectra because of line broadening | |
| Glycine [19] : 68 | 3.55 | - Inhibitory neurotransmitter
- Forms significant fraction of collagen
| None | - Elevated levels in infants with hyperglycinemia and patients with brain tumors [20]
|
| Glycogen [19] : 68–69 | 3.83 | - Form of energy storage
- Important role in systemic glucose metabolism
| Routinely observed in 13C NMR, but remains elusive in 1H NMR | - Altered levels in diabetes mellitus
|
| Histidine [19] : 59–70 | 7.10, 7.80 | - Precursor for histamine and carnosine biosynthesis
| Establish intracellular pH in 1H NMR | - Elevated levels in hepatic encephalopathy and histidinemia
|
| Homocarnosine [19] : 70 | 7.10, 8.10, 3.00 - 4.50 | - Associated with epileptic seizure control
| Good choice for in vivo pH monitoring Because of the overlap between GABA and Homocarnosine resonances, the GABA H-4 resonance at 3.01 ppm is the "total GABA" representing the sum of GABA and homocarnosine | - Elevated levels seen in antiepileptic drugs, like gabapentin
- Elevated levels in brain and CSF are associated with homocarnosinase
|
| β-Hydroxybutyrate (BHB) [19] : 70–71 | 1.19 | - Alternate substrate for metabolism, typically under conditions of long fasting or high fat diets
- Maintain acetoacetyl-CoA and acetyl-CoA for the synthesis of cholesterol, fatty acids, and complex lipids
| None | - Elevated levels believed to control seizures in childhood epilepsy
|
| 2-Hydroxyglutarate (2HG) [19] : 71–72 | 1.90 | - Oncometabolite (cancer-causing)
- Part of butanoate metabolic pathway
| None | - Elevated levels in gliomas
|
| myo-Inositol (mI) [19] : 72–73 | 3.52 | - Exact function is unknown
- Osmotic regulation in kidney
- Biochemical relationship to messenger-inositol polyphosphate
| None | - Altered levels in patients with mild cognitive impairment, Alzheimer's disease, and brain injury
|
| scyllo-Inositol (sI) [19] : 72–73 | 3.34 | - Reverse memory deficits
- Reduce development of amyloid-beta (Aβ) plaques
| None | - Elevated levels during chronic alcoholism
|
| Lactate (Lac) [19] : 73–74 | 1.31 | - End product of anaerobic glycolysis
- Links astroglial glucose uptake and metabolism to neuronal neurotransmitter cycling in astroglial-neuronal lactate shuttle hypothesis (ANLS)
| None | - Elevated levels seen in hyperventilation, tumors, ischemic stroke, hypoxia
|
| Lipids [19] : 87 | 0.9 - 1.5 | - Intramyocellular lipids represents a pool that displays active turnover and metabolism, for example, during exercise
- Extramyocellular lipids represent an inert pool that lies between muscle fibers
| High abundance of lipids is one of main reasons 1H NMR outside the brain has seen limited applications | - Elevated levels seen in necrosis
|
| Macromolecules [19] : 74–76 | 0.93 (MM1), 1.24 (MM2), 1.43 (MM3), 1.72 (MM4), 2.05 (MM5), 2.29 (MM6), 3.00 (MM7), 3.20 (MM8), 3.8 - 4.0 (MM9), 4.3 (MM10) | - Assignment to specific proteins is essentially impossible, but individual resonances can be assigned to amino acids
- MM1: Leucine, Isoleucine, Valine
- MM2 and MM3: Threonine and Alanine
- MM4 and MM7: Lysine and Arginine
- MM5 and MM6: Glutamate and Glutamine
- MM8-MM10: Not very well defined to know
| Significant fraction of observed signal is macromolecular resonances underlying the rest of metabolites Short T2 relaxation time constants effectively eliminate macromolecular resonances from long-echo-time 1H NMR spectra Difference in T1 relaxations between metabolites and macromolecules is used to reduce contribution from extracranial lipid signal | - Alternations in macromolecular spectrum observed in stroke, tumors, multiple sclerosis, and aging
|
| Nicotinamid Adenine Dinucleotide (NAD+) [19] : 76 | 9.00 | - Coenzyme for electron-transfer enzymes
- Substrate for ADP-ribose transferases, poly (ADP-ribose) polymerases, cADP-ribose synthases, and sirtuins
- Involved in gene expression and repair, calcium mobilization, metabolism, aging, cancer, cell metabolism, and the timing of metabolism via the circadian rhythm
| 31P NMR allows detection of both NAD+ and NADH, while 1H NMR does not allow detection for NADH | |
| Phenylalanine [19] : 76–77 | 7.30 - 7.45 | - Precursor for the amino acid tyrosine which is used for catchelcolamine (dopamine, epinephrine, and norepinephrine) synthesis
| None | - Elevated levels in phenylketonuria (PKU)
- Reduced levels in aging
|
| Pyruvate [19] : 77–78 | 2.36 | - Converted into acetyl-coenzyme A
- Participates in the anaplerotic reaction to replenish TCA cycle intermediates
- Neuroprotective properties in stroke
| Only FDA-approved compound for hyperpolarized 13C NMR | - Altered levels in cystic lesions and neonatal pyruvate dehydrogenase deficiency
|
| Serine [19] : 78 | 3.80 - 4.00 | - Participates in biosynthesis of purines, pyridines, cysteine, glycine, 3-phosphoglycerate, and other proteins
| None | - Elevated levels in patients with Alzheimer's
|
| Taurine (Tau) [19] : 79–80 | 3.25, 3.42 | - Exact function not known
- Osmoregulator
- Modulator of neurotransmitters
| None | |
| Threonine (Thr) [19] : 80 | 1.32 | | None | - Supplement to help alleviate anxiety and some cases of depression
|
| Tryptophan (Trp) [19] : 80 | 7.20, 7.28 | - Necessary for production of serotonin, melatonin, vitamin B3 (niacin), and NAD+
| None | - Elevated levels seen in hepatic encephalopathy
- Treatment for mild insomnia
- Antidepressant
|
| Tyrosine (Tyr) [19] : 81 | 6.89 - 7.19 | - Precursor to the neurotransmitters epinephrine, norepinephrine, and dopamine as well as the thyroid hormones thyroxine and triiodothyronine
- Converted to DOPA by tyrosine dehydroxylase
- Key role in signal transduction
| None | - Elevated levels in hepatic encephalopathy
- Reduced levels seen with aging
|
| Water [19] : 81–82 | 4.80 | | Internal concentration referencing Water chemical shift used to detect temperature changes noninvasively in vivo | - Water content only changes moderately with different pathologies
|