DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages. This can eventually lead to malignant tumors, or cancer as per the two hit hypothesis.

Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair(HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber.

Mycobacterium smegmatis is an acid-fast bacterial species in the phylum Actinomycetota and the genus Mycobacterium. It is 3.0 to 5.0 µm long with a bacillus shape and can be stained by Ziehl–Neelsen method and the auramine-rhodamine fluorescent method. It was first reported in November 1884 by Lustgarten, who found a bacillus with the staining appearance of tubercle bacilli in syphilitic chancres. Subsequent to this, Alvarez and Tavel found organisms similar to that described by Lustgarten also in normal genital secretions (smegma). This organism was later named M. smegmatis.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids.

Werner syndrome ATP-dependent helicase, also known as DNA helicase, RecQ-like type 3, is an enzyme that in humans is encoded by the WRN gene. WRN is a member of the RecQ Helicase family. Helicase enzymes generally unwind and separate double-stranded DNA. These activities are necessary before DNA can be copied in preparation for cell division. Helicase enzymes are also critical for making a blueprint of a gene for protein production, a process called transcription. Further evidence suggests that Werner protein plays a critical role in repairing DNA. Overall, this protein helps maintain the structure and integrity of a person's DNA.

Serine/threonine-protein kinase ATR also known as ataxia telangiectasia and Rad3-related protein (ATR) or FRAP-related protein 1 (FRP1) is an enzyme that, in humans, is encoded by the ATR gene. It is a large kinase of about 301.66 kDa. ATR belongs to the phosphatidylinositol 3-kinase-related kinase protein family. ATR is activated in response to single strand breaks, and works with ATM to ensure genome integrity.

DNA repair protein XRCC4 also known as X-ray repair cross-complementing protein 4 or XRCC4 is a protein that in humans is encoded by the XRCC4 gene. In addition to humans, the XRCC4 protein is also expressed in many other metazoans, fungi and in plants. The X-ray repair cross-complementing protein 4 is one of several core proteins involved in the non-homologous end joining (NHEJ) pathway to repair DNA double strand breaks (DSBs).

Ku70 is a protein that, in humans, is encoded by the XRCC6 gene.

DNA-dependent protein kinase, catalytic subunit, also known as DNA-PKcs, is an enzyme that in humans is encoded by the gene designated as PRKDC or XRCC7. DNA-PKcs belongs to the phosphatidylinositol 3-kinase-related kinase protein family. The DNA-Pkcs protein is a serine/threonine protein kinase comprising a single polypeptide chain of 4,128 amino acids.

Ku80 is a protein that, in humans, is encoded by the XRCC5 gene. Together, Ku70 and Ku80 make up the Ku heterodimer, which binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. It is also required for V(D)J recombination, which utilizes the NHEJ pathway to promote antigen diversity in the mammalian immune system.

DNA polymerase mu is a polymerase enzyme found in eukaryotes. In humans, this protein is encoded by the POLM gene.
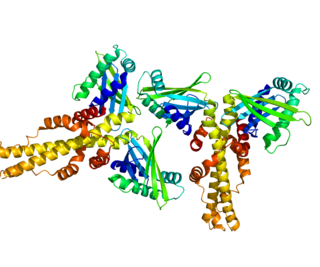
Non-homologous end-joining factor 1 (NHEJ1), also known as Cernunnos or XRCC4-like factor (XLF), is a protein that in humans is encoded by the NHEJ1 gene. XLF was originally discovered as the protein mutated in five patients with growth retardation, microcephaly, and immunodeficiency. The protein is required for the non-homologous end joining (NHEJ) pathway of DNA repair. Patients with XLF mutations also have immunodeficiency due to a defect in V(D)J recombination, which uses NHEJ to generate diversity in the antibody repertoire of the immune system. XLF interacts with DNA ligase IV and XRCC4 and is thought to be involved in the end-bridging or ligation steps of NHEJ. The yeast homolog of XLF is Nej1.

Histone-lysine N-methyltransferase SETMAR is an enzyme that in humans is encoded by the SETMAR gene.
Microhomology-mediated end joining (MMEJ), also known as alternative nonhomologous end-joining (Alt-NHEJ) is one of the pathways for repairing double-strand breaks in DNA. As reviewed by McVey and Lee, the foremost distinguishing property of MMEJ is the use of microhomologous sequences during the alignment of broken ends before joining, thereby resulting in deletions flanking the original break. MMEJ is frequently associated with chromosome abnormalities such as deletions, translocations, inversions and other complex rearrangements.
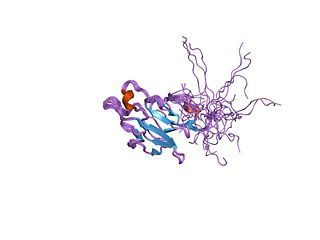
In molecular biology, the forkhead-associated domain is a phosphopeptide recognition domain found in many regulatory proteins. It displays specificity for phosphothreonine-containing epitopes but will also recognise phosphotyrosine with relatively high affinity. It spans approximately 80-100 amino acid residues folded into an 11-stranded beta sandwich, which sometimes contains small helical insertions between the loops connecting the strands.
Shelterin is a protein complex known to protect telomeres in many eukaryotes from DNA repair mechanisms, as well as to regulate telomerase activity. In mammals and other vertebrates, telomeric DNA consists of repeating double-stranded 5'-TTAGGG-3' (G-strand) sequences along with the 3'-AATCCC-5' (C-strand) complement, ending with a 50-400 nucleotide 3' (G-strand) overhang. Much of the final double-stranded portion of the telomere forms a T-loop (Telomere-loop) that is invaded by the 3' (G-strand) overhang to form a small D-loop (Displacement-loop).

Cell cycle regulator of non-homologous end joining is a protein that in humans is encoded by the CYREN gene.
Telomeres, the caps on the ends of eukaryotic chromosomes, play critical roles in cellular aging and cancer. An important facet to how telomeres function in these roles is their involvement in cell cycle regulation.

DNA end resection, also called 5′–3′ degradation, is a biochemical process where the blunt end of a section of double-stranded DNA (dsDNA) is modified by cutting away some nucleotides from the 5' end to produce a 3' single-stranded sequence. The presence of a section of single-stranded DNA (ssDNA) allows the broken end of the DNA to line up accurately with a matching sequence, so that it can be accurately repaired.

A double-strand break repair model refers to the various models of pathways that cells undertake to repair double strand-breaks (DSB). DSB repair is an important cellular process, as the accumulation of unrepaired DSB could lead to chromosomal rearrangements, tumorigenesis or even cell death. In human cells, there are two main DSB repair mechanisms: Homologous recombination (HR) and non-homologous end joining (NHEJ). HR relies on undamaged template DNA as reference to repair the DSB, resulting in the restoration of the original sequence. NHEJ modifies and ligates the damaged ends regardless of homology. In terms of DSB repair pathway choice, most mammalian cells appear to favor NHEJ rather than HR. This is because the employment of HR may lead to gene deletion or amplification in cells which contains repetitive sequences. In terms of repair models in the cell cycle, HR is only possible during the S and G2 phases, while NHEJ can occur throughout whole process. These repair pathways are all regulated by the overarching DNA damage response mechanism. Besides HR and NHEJ, there are also other repair models which exists in cells. Some are categorized under HR, such as synthesis-dependent strain annealing, break-induced replication, and single-strand annealing; while others are an entirely alternate repair model, namely, the pathway microhomology-mediated end joining (MMEJ).