
DNA adenine methylase, (Dam methylase) (also site-specific DNA-methyltransferase (adenine-specific), EC 2.1.1.72, modification methylase, restriction-modification system) is an enzyme that adds a methyl group to the adenine of the sequence 5'-GATC-3' in newly synthesized DNA. Immediately after DNA synthesis, the daughter strand remains unmethylated for a short time. It is an orphan methyltransferase that is not part of a restriction-modification system and regulates gene expression. This enzyme catalyses the following chemical reaction
In enzymology, a tRNA guanosine-2'-O-methyltransferase is an enzyme that catalyzes the chemical reaction
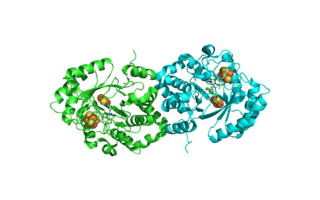
Biotin synthase (BioB) is an enzyme that catalyzes the conversion of dethiobiotin (DTB) to biotin; this is the final step in the biotin biosynthetic pathway. Biotin, also known as vitamin B7, is a cofactor used in carboxylation, decarboxylation, and transcarboxylation reactions in many organisms including humans. Biotin synthase is an S-Adenosylmethionine (SAM) dependent enzyme that employs a radical mechanism to thiolate dethiobiotin, thus converting it to biotin.

Cobalamin biosynthesis is the process by which bacteria and archea make cobalamin, vitamin B12. Many steps are involved in converting aminolevulinic acid via uroporphyrinogen III and adenosylcobyric acid to the final forms in which it is used by enzymes in both the producing organisms and other species, including humans who acquire it through their diet.
Radical SAM is a designation for a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. rSAMs comprise the largest superfamily of metal-containing enzymes.

Uroporphyrinogen-III C-methyltransferase, uroporphyrinogen methyltransferase, uroporphyrinogen-III methyltransferase, adenosylmethionine-uroporphyrinogen III methyltransferase, S-adenosyl-L-methionine-dependent uroporphyrinogen III methylase, uroporphyrinogen-III methylase, SirA, CysG, CobA, uroporphyrin-III C-methyltransferase, S-adenosyl-L-methionine:uroporphyrin-III C-methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:uroporphyrinogen-III C-methyltransferase. This enzyme catalyses the following chemical reaction
Demethylmenaquinone methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:demethylmenaquinone methyltransferase. This enzyme catalyses the following chemical reaction
Methyl halide transferase is an enzyme with systematic name S-adenosylmethionine:iodide methyltransferase. This enzyme catalyses the following chemical reaction
23S rRNA (uridine2552-2'-O)-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (uridine2552-2'-O-)-methyltransferase. This enzyme catalyses the following chemical reaction
16S rRNA (guanine1207-N2)-methyltransferase (EC 2.1.1.172, m2G1207 methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:16S rRNA (guanine1207-N2)-methyltransferase. This enzyme catalyses the following chemical reaction
16S rRNA (adenine1408-N1)-methyltransferase (EC 2.1.1.180, kanamycin-apramycin resistance methylase, 16S rRNA:m1A1408 methyltransferase, KamB, NpmA, 16S rRNA m1A1408 methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:16S rRNA (adenine1408-N1)-methyltransferase. This enzyme catalyses the following chemical reaction
16S rRNA (adenine1518-N6/adenine1519-N6)-dimethyltransferase (EC 2.1.1.182, S-adenosylmethionine-6-N',N'-adenosyl (rRNA) dimethyltransferase, KsgA, ksgA methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:16S rRNA (adenine1518-N6/adenine1519-N6)-dimethyltransferase. This enzyme catalyses the following chemical reaction
23S rRNA (guanosine2251-2'-O)-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (guanosine2251-2'-O-)-methyltransferase. This enzyme catalyses the following chemical reaction
23S rRNA (guanine745-N1)-methyltransferase (EC 2.1.1.187, Rlma(I), Rlma1, 23S rRNA m1G745 methyltransferase, YebH, RlmAI methyltransferase, ribosomal RNA(m1G)-methylase, rRNA(m1G)methylase, RrmA, 23S rRNA:m1G745 methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (guanine745-N1)-methyltransferase. This enzyme catalyses the following chemical reaction
23S rRNA (uracil1939-C5)-methyltransferase (EC 2.1.1.190, RumA, RNA uridine methyltransferase A, YgcA) is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (uracil1939-C5)-methyltransferase. This enzyme catalyses the following chemical reaction
TRNA (cytidine32/uridine32-2'-O)-methyltransferase (EC 2.1.1.200, YfhQ, tRNA:Cm32/Um32 methyltransferase, TrMet(Xm32), TrmJ) is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (cytidine32/uridine32-2'-O)-methyltransferase. This enzyme catalyses the following chemical reaction
tRNA (cytidine34-2'-O)-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (cytidine34/5-carboxymethylaminomethyluridine34-2'-O)-methyltransferase. This enzyme catalyses the following chemical reaction
tRNA (guanine37-N1)-methyltransferase (EC 2.1.1.228, TrmD, tRNA (m1G37) methyltransferase, transfer RNA (m1G37) methyltransferase, Trm5p, TRMT5, tRNA-(N1G37) methyltransferase, MJ0883 (gene)) is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (guanine37-N1)-methyltransferase. This enzyme catalyses the following chemical reaction
Erythromycin 3''-O-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:erythromycin C 3''-O-methyltransferase. This enzyme catalyses the following chemical reaction
Tellurite methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:tellurite methyltransferase. This enzyme catalyses the following chemical reaction




