
Genetic programming (GP) is an evolutionary algorithm, an artificial intelligence technique mimicking natural evolution, which operates on a population of programs. It applies the genetic operators selection according to a predefined fitness measure, mutation and crossover.

In computer science and operations research, a genetic algorithm (GA) is a metaheuristic inspired by the process of natural selection that belongs to the larger class of evolutionary algorithms (EA). Genetic algorithms are commonly used to generate high-quality solutions to optimization and search problems via biologically inspired operators such as selection, crossover, and mutation. Some examples of GA applications include optimizing decision trees for better performance, solving sudoku puzzles, hyperparameter optimization, and causal inference.

Evolutionary algorithms (EA) reproduce essential elements of the biological evolution in a computer algorithm in order to solve “difficult” problems, at least approximately, for which no exact or satisfactory solution methods are known. They belong to the class of metaheuristics and are a subset of evolutionary computation, which itself is part of the field of computational intelligence. The mechanisms of biological evolution that an EA mainly imitates are reproduction, mutation, recombination and selection. Candidate solutions to the optimization problem play the role of individuals in a population, and the fitness function determines the quality of the solutions (see also loss function). Evolution of the population then takes place after the repeated application of the above operators.

Evolutionary computation from computer science is a family of algorithms for global optimization inspired by biological evolution, and the subfield of artificial intelligence and soft computing studying these algorithms. In technical terms, they are a family of population-based trial and error problem solvers with a metaheuristic or stochastic optimization character.

A genetic operator is an operator used in evolutionary algorithms to guide the algorithm towards a solution to a given problem. There are three main types of operators, which must work in conjunction with one another in order for the algorithm to be successful. Genetic operators are used to create and maintain genetic diversity, combine existing solutions into new solutions (crossover) and select between solutions (selection). In his book discussing the use of genetic programming for the optimization of complex problems, computer scientist John Koza has also identified an 'inversion' or 'permutation' operator; however, the effectiveness of this operator has never been conclusively demonstrated and this operator is rarely discussed.

A fitness function is a particular type of objective or cost function that is used to summarize, as a single figure of merit, how close a given candidate solution is to achieving the set aims. It is an important component of evolutionary algorithms (EA), such as genetic programming, evolution strategies or genetic algorithms. An EA is a metaheuristic that reproduces the basic principles of biological evolution as a computer algorithm in order to solve challenging optimization or planning tasks, at least approximately. For this purpose, many candidate solutions are generated, which are evaluated using a fitness function in order to guide the evolutionary development towards the desired goal. Similar quality functions are also used in other metaheuristics, such as ant colony optimization or particle swarm optimization.

Crossover in evolutionary algorithms and evolutionary computation, also called recombination, is a genetic operator used to combine the genetic information of two parents to generate new offspring. It is one way to stochastically generate new solutions from an existing population, and is analogous to the crossover that happens during sexual reproduction in biology. New solutions can also be generated by cloning an existing solution, which is analogous to asexual reproduction. Newly generated solutions may be mutated before being added to the population. The aim of recombination is to transfer good characteristics from two different parents to one child.

A chromosome or genotype in evolutionary algorithms (EA) is a set of parameters which define a proposed solution of the problem that the evolutionary algorithm is trying to solve. The set of all solutions, also called individuals according to the biological model, is known as the population. The genome of an individual consists of one, more rarely of several, chromosomes and corresponds to the genetic representation of the task to be solved. A chromosome is composed of a set of genes, where a gene consists of one or more semantically connected parameters, which are often also called decision variables. They determine one or more phenotypic characteristics of the individual or at least have an influence on them. In the basic form of genetic algorithms, the chromosome is represented as a binary string, while in later variants and in EAs in general, a wide variety of other data structures are used.
Metaheuristic in computer science and mathematical optimization is a higher-level procedure or heuristic designed to find, generate, tune, or select a heuristic that may provide a sufficiently good solution to an optimization problem or a machine learning problem, especially with incomplete or imperfect information or limited computation capacity. Metaheuristics sample a subset of solutions which is otherwise too large to be completely enumerated or otherwise explored. Metaheuristics may make relatively few assumptions about the optimization problem being solved and so may be usable for a variety of problems. Their use is always of interest when exact or other (approximate) methods are not available or are not expedient, either because the calculation time is too long or because, for example, the solution provided is too imprecise.

Evolution strategy (ES) from computer science is a subclass of evolutionary algorithms, which serves as an optimization technique. It uses the major genetic operators mutation, recombination and selection of parents.

Gene expression programming (GEP) in computer programming is an evolutionary algorithm that creates computer programs or models. These computer programs are complex tree structures that learn and adapt by changing their sizes, shapes, and composition, much like a living organism. And like living organisms, the computer programs of GEP are also encoded in simple linear chromosomes of fixed length. Thus, GEP is a genotype–phenotype system, benefiting from a simple genome to keep and transmit the genetic information and a complex phenotype to explore the environment and adapt to it.

Selection is a genetic operator in a evolutionary algorithm in which individual genomes are chosen from a population for later breeding. Selection mechanisms are also used to choose candidate solutions (individuals) for the next generation. Retaining the best individuals in a generation unchanged in the next generation, is called elitism or elitist selection. It is a successful (slight) variant of the general process of constructing a new population.

Premature convergence is an unwanted effect in evolutionary algorithms (EA), a metaheuristic that mimics the basic principles of biological evolution as a computer algorithm for solving an optimization problem. The effect means that the population of an EA has converged too early, resulting in being suboptimal. In this context, the parental solutions, through the aid of genetic operators, are not able to generate offspring that are superior to, or outperform, their parents. Premature convergence is a common problem found in evolutionary algorithms, as it leads to a loss, or convergence of, a large number of alleles, subsequently making it very difficult to search for a specific gene in which the alleles were present. An allele is considered lost if, in a population, a gene is present, where all individuals are sharing the same value for that particular gene. An allele is, as defined by De Jong, considered to be a converged allele, when 95% of a population share the same value for a certain gene.

Genetic distance is a measure of the genetic divergence between species or between populations within a species, whether the distance measures time from common ancestor or degree of differentiation. Populations with many similar alleles have small genetic distances. This indicates that they are closely related and have a recent common ancestor.
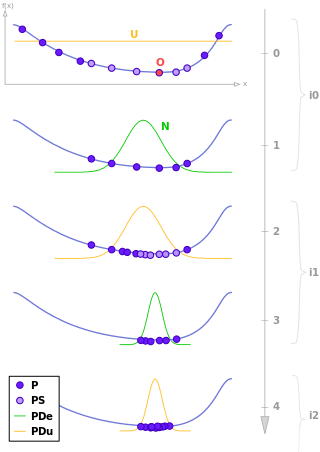
Estimation of distribution algorithms (EDAs), sometimes called probabilistic model-building genetic algorithms (PMBGAs), are stochastic optimization methods that guide the search for the optimum by building and sampling explicit probabilistic models of promising candidate solutions. Optimization is viewed as a series of incremental updates of a probabilistic model, starting with the model encoding an uninformative prior over admissible solutions and ending with the model that generates only the global optima.

A point accepted mutation — also known as a PAM — is the replacement of a single amino acid in the primary structure of a protein with another single amino acid, which is accepted by the processes of natural selection. This definition does not include all point mutations in the DNA of an organism. In particular, silent mutations are not point accepted mutations, nor are mutations that are lethal or that are rejected by natural selection in other ways.

In computer programming, genetic representation is a way of presenting solutions/individuals in evolutionary computation methods. The term encompasses both the concrete data structures and data types used to realize the genetic material of the candidate solutions in the form of a genome, and the relationships between search space and problem space. In the simplest case, the search space corresponds to the problem space. The choice of problem representation is tied to the choice of genetic operators, both of which have a decisive effect on the efficiency of the optimization. Genetic representation can encode appearance, behavior, physical qualities of individuals. Difference in genetic representations is one of the major criteria drawing a line between known classes of evolutionary computation.
Biogeography-based optimization (BBO) is an evolutionary algorithm (EA) that optimizes a function by stochastically and iteratively improving candidate solutions with regard to a given measure of quality, or fitness function. BBO belongs to the class of metaheuristics since it includes many variations, and since it does not make any assumptions about the problem and can therefore be applied to a wide class of problems.

The Fly Algorithm is a computational method within the field of evolutionary algorithms, designed for direct exploration of 3D spaces in applications such as computer stereo vision, robotics, and medical imaging. Unlike traditional image-based stereovision, which relies on matching features to construct 3D information, the Fly Algorithm operates by generating a 3D representation directly from random points, termed "flies." Each fly is a coordinate in 3D space, evaluated for its accuracy by comparing its projections in a scene. By iteratively refining the positions of flies based on fitness criteria, the algorithm can construct an optimized spatial representation. The Fly Algorithm has expanded into various fields, including applications in digital art, where it is used to generate complex visual patterns.

Genotypic and phenotypic repair are optional components of an evolutionary algorithm (EA). An EA reproduces essential elements of biological evolution as a computer algorithm in order to solve demanding optimization or planning tasks, at least approximately. A candidate solution is represented by a - usually linear - data structure that plays the role of an individual's chromosome. New solution candidates are generated by mutation and crossover operators following the example of biology. These offspring may be defective, which is corrected or compensated for by genotypic or phenotypic repair.











































