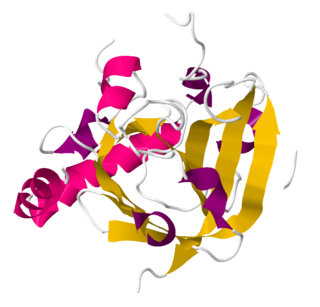Lipoic acid (LA), also known as α-lipoic acid, alpha-lipoic acid (ALA) and thioctic acid, is an organosulfur compound derived from caprylic acid (octanoic acid). ALA is made in animals normally, and is essential for aerobic metabolism. It is also manufactured and is available as a dietary supplement in some countries where it is marketed as an antioxidant, and is available as a pharmaceutical drug in other countries. Lipoate is the conjugate base of lipoic acid, and the most prevalent form of LA under physiological conditions. Only the (R)-(+)-enantiomer (RLA) exists in nature and is essential for aerobic metabolism because RLA is an essential cofactor of many enzyme complexes.

Lysins, also known as endolysins or murein hydrolases, are hydrolytic enzymes produced by bacteriophages in order to cleave the host's cell wall during the final stage of the lytic cycle. Lysins are highly evolved enzymes that are able to target one of the five bonds in peptidoglycan (murein), the main component of bacterial cell walls, which allows the release of progeny virions from the lysed cell. Cell-wall-containing Archaea are also lysed by specialized pseudomurein-cleaving lysins, while most archaeal viruses employ alternative mechanisms. Similarly, not all bacteriophages synthesize lysins: some small single-stranded DNA and RNA phages produce membrane proteins that activate the host's autolytic mechanisms such as autolysins.

Glycine decarboxylase also known as glycine cleavage system P protein or glycine dehydrogenase is an enzyme that in humans is encoded by the GLDC gene.

Lipoyl synthase is an enzyme that belongs to the radical SAM (S-adenosyl methionine) family. Within the radical SAM superfamily, lipoyl synthase is in a sub-family of enzymes that catalyze sulfur insertion reactions. The enzymes in this subfamily differ from general radical SAM enzymes, as they contain two 4Fe-4S clusters. From these clusters, the enzymes obtain the sulfur groups that will be transferred onto the corresponding substrates. This particular enzyme participates in the final step of lipoic acid metabolism, transferring two sulfur atoms from its 4Fe-4S cluster onto the protein N6-(octanoyl)lysine through radical generation. This enzyme is usually localized to the mitochondria. Two organisms that have been extensively studied with regards to this enzyme are Escherichia coli and Mycobacterium tuberculosis. It is currently being studied in other organisms including yeast, plants, and humans.
In enzymology, a lipoyl(octanoyl) transferase (EC 2.3.1.181) is an enzyme that catalyzes the chemical reaction

Glycine cleavage system H protein, mitochondrial is a protein that in humans is encoded by the GCSH gene. Degradation of glycine is brought about by the glycine cleavage system (GCS), which is composed of 4 protein components: P protein, H protein, T protein, and L protein. The H protein shuttles the methylamine group of glycine from the P protein to the T protein. The protein encoded by GCSH gene is the H protein, which transfers the methylamine group of glycine from the P protein to the T protein. Defects in this gene are a cause of nonketotic hyperglycinemia (NKH). Two transcript variants, one protein-coding and the other probably not protein-coding, have been found for this gene. Also, several transcribed and non-transcribed pseudogenes of this gene exist throughout the genome.
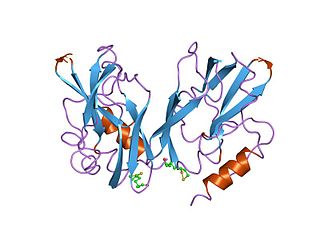
The glycine cleavage system (GCS) is also known as the glycine decarboxylase complex or GDC. The system is a series of enzymes that are triggered in response to high concentrations of the amino acid glycine. The same set of enzymes is sometimes referred to as glycine synthase when it runs in the reverse direction to form glycine. The glycine cleavage system is composed of four proteins: the T-protein, P-protein, L-protein, and H-protein. They do not form a stable complex, so it is more appropriate to call it a "system" instead of a "complex". The H-protein is responsible for interacting with the three other proteins and acts as a shuttle for some of the intermediate products in glycine decarboxylation. In both animals and plants, the glycine cleavage system is loosely attached to the inner membrane of the mitochondria. Mutations in this enzymatic system are linked with glycine encephalopathy.
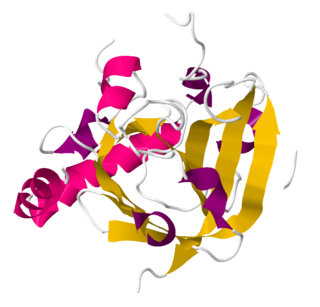
Sortase refers to a group of prokaryotic enzymes that modify surface proteins by recognizing and cleaving a carboxyl-terminal sorting signal. For most substrates of sortase enzymes, the recognition signal consists of the motif LPXTG (Leu-Pro-any-Thr-Gly), then a highly hydrophobic transmembrane sequence, followed by a cluster of basic residues such as arginine. Cleavage occurs between the Thr and Gly, with transient attachment through the Thr residue to the active site Cys residue, followed by transpeptidation that attaches the protein covalently to cell wall components. Sortases occur in almost all Gram-positive bacteria and the occasional Gram-negative bacterium or Archaea, where cell wall LPXTG-mediated decoration has not been reported. Although sortase A, the "housekeeping" sortase, typically acts on many protein targets, other forms of sortase recognize variant forms of the cleavage motif, or catalyze the assembly of pilins into pili.

In molecular biology, the Cofactor transferase family is a family of protein domains that includes biotin protein ligases, lipoate-protein ligases A, octanoyl-(acyl carrier protein):protein N-octanoyltransferases, and lipoyl-protein:protein N-lipoyltransferases. The metabolism of the cofactors Biotin and lipoic acid share this family. They also share the target modification domain, and the sulfur insertion enzyme.
In molecular biology, the SR1 RNA is a small RNA (sRNA) produced by species of Bacillus and closely related bacteria. It is a dual-function RNA which acts both as a protein-coding RNA and as a regulatory sRNA.
Acyl-homoserine-lactone synthase is an enzyme with systematic name acyl-(acyl-carrier protein):S-adenosyl-L-methionine acyltranserase . This enzyme catalyses the following chemical reaction
Lipoyl amidotransferase (EC 2.3.1.200, LipL (gene)) is an enzyme with systematic name (glycine cleavage system H)-N6-lipoyl-L-lysine:(lipoyl-carrier protein)-N6-L-lysine lipoyltransferase. This enzyme catalyses the following chemical reaction
Lipid II:glycine glycyltransferase (EC 2.3.2.16, N-acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-D-alanyl-D-alanine-diphosphoundecaprenyl-N-acetylglucosamine:N6-glycine transferase, femX (gene)) is an enzyme with systematic name alanyl-D-alanine-diphospho-ditrans, octacis-undecaprenyl-N-acetylglucosamine:glycine N6-glycyltransferase. This enzyme catalyses the following chemical reaction
N-acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-(N6-glycyl)-D-alanyl-D-alanine-diphosphoundecaprenyl-N-acetylglucosamine:glycine glycyltransferase (EC 2.3.2.17, femA (gene)) is an enzyme with systematic name N-acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-(N6-glycyl)-D-alanyl-D-alanine-ditrans,octacis-diphosphoundecaprenyl-N-acetylglucosamine:glycine glycyltransferase. This enzyme catalyses the following chemical reaction
N-acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-(N6-triglycine)-D-alanyl-D-alanine-diphosphoundecaprenyl-N-acetylglucosamine:glycine glycyltransferase (EC 2.3.2.18, femB (gene)) is an enzyme with systematic name N-acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-(N6-triglycine)-D-alanyl-D-alanine-ditrans,octacis-diphosphoundecaprenyl-N-acetylglucosamine:glycine glycyltransferase. This enzyme catalyses the following chemical reaction
Lipoate–protein ligase (EC 2.7.7.63, LplA, lipoate protein ligase, lipoate–protein ligase A, LPL, LPL-B) is an enzyme with systematic name ATP:lipoate adenylyltransferase. This enzyme catalyses the following chemical reaction
The enzyme Pimelyl-[acyl-carrier protein] methyl ester esterase (EC 3.1.1.85, BioH; systematic name pimelyl-[acyl-carrier protein] methyl ester hydrolase catalyses the reaction
SpoIVB peptidase is an enzyme. This enzyme catalyses the following chemical reaction
Lysine Exporters are a superfamily of transmembrane proteins which export amino acids, lipids and heavy metal ions. They provide ionic homeostasis, play a role in cell envelope assembly, and protect from excessive concentrations of heavy metals in cytoplasm. The superfamily was named based on the early discovery of the LysE carrier protein of Corynebacterium glutamicum.
The gluconate:H+ symporter (GntP) family (TC# 2.A.8) is a family of transport proteins belonging to the ion transporter (IT) superfamily. Members of the GntP family include known gluconate permeases of E. coli and Bacillus species such as the D-Gluconate:H+ symporter of Bacillus subtillus (GntP; TC# 2.A.8.1.1) and the D-fructuronate/D-gluconate:H+ symporter of E. coli (GntP; TC# 2.A.8.1.3). A representative list of proteins belonging to the GntP family can be found in the Transporter Classification Database.