
Proteasome subunit alpha type-3 also known as macropain subunit C8 and proteasome component C8 is a protein that in humans is encoded by the PSMA3 gene. This protein is one of the 17 essential subunits that contributes to the complete assembly of 20S proteasome complex.

Proteasome subunit alpha type-4 also known as macropain subunit C9, proteasome component C9, and 20S proteasome subunit alpha-3 is a protein that in humans is encoded by the PSMA4 gene. This protein is one of the 17 essential subunits that contributes to the complete assembly of 20S proteasome complex.
Proteasome subunit beta type-4 also known as 20S proteasome subunit beta-7 is a protein that in humans is encoded by the PSMB4 gene.

26S proteasome non-ATPase regulatory subunit 13 is an enzyme that in humans is encoded by the PSMD13 gene.

Proteasome subunit beta type-10 as known as 20S proteasome subunit beta-2i is a protein that in humans is encoded by the PSMB10 gene.
Proteasome subunit beta type-2 also known as 20S proteasome subunit beta-4 is a protein that in humans is encoded by the PSMB2 gene. This protein is one of the 17 essential subunits that contributes to the complete assembly of 20S proteasome complex. In particular, proteasome subunit beta type-2, along with other beta subunits, assemble into two heptameric rings and subsequently a proteolytic chamber for substrate degradation. The eukaryotic proteasome recognized degradable proteins, including damaged proteins for protein quality control purpose or key regulatory protein components for dynamic biological processes. An essential function of a modified proteasome, the immunoproteasome, is the processing of class I MHC peptides.

Proteasome subunit beta type-3, also known as 20S proteasome subunit beta-3, is a protein that in humans is encoded by the PSMB3 gene. This protein is one of the 17 essential subunits that contribute to the complete assembly of the 20S proteasome complex. In particular, proteasome subunit beta type-2, along with other beta subunits, assemble into two heptameric rings and subsequently a proteolytic chamber for substrate degradation. The eukaryotic proteasome recognizes degradable proteins, including damaged proteins for protein quality control purpose or key regulatory protein components for dynamic biological processes.

Proteasome subunit beta type-8 as known as 20S proteasome subunit beta-5i is a protein that in humans is encoded by the PSMB8 gene. This protein is one of the 17 essential subunits that contributes to the complete assembly of 20S proteasome complex. In particular, proteasome subunit beta type-5, along with other beta subunits, assemble into two heptameric rings and subsequently a proteolytic chamber for substrate degradation. This protein contains "Chymotrypsin-like" activity and is capable of cleaving after large hydrophobic residues of peptide. The eukaryotic proteasome recognized degradable proteins, including damaged proteins for protein quality control purpose or key regulatory protein components for dynamic biological processes. The constitutive subunit beta1, beta2, and beta 5 can be replaced by their inducible counterparts beta1i, 2i, and 5i when cells are under the treatment of interferon-γ. The resulting proteasome complex becomes the so-called immunoproteasome. An essential function of the modified proteasome complex, the immunoproteasome, is the processing of numerous MHC class-I restricted T cell epitopes.

Proteasome subunit beta type-9 as known as 20S proteasome subunit beta-1i is a protein that in humans is encoded by the PSMB9 gene.

Proteasome subunit alpha type-6 is a protein that in humans is encoded by the PSMA6 gene. This protein is one of the 17 essential subunits that contributes to the complete assembly of 20S proteasome complex.

Proteasome subunit alpha type-1 is a protein that in humans is encoded by the PSMA1 gene. This protein is one of the 17 essential subunits that contributes to the complete assembly of 20S proteasome complex.

Proteasome subunit alpha type-7 also known as 20S proteasome subunit alpha-4 is a protein that in humans is encoded by the PSMA7 gene. This protein is one of the 17 essential subunits that contributes to the complete assembly of 20S proteasome complex.
Proteasome subunit beta type-5 as known as 20S proteasome subunit beta-5 is a protein that in humans is encoded by the PSMB5 gene. This protein is one of the 17 essential subunits that contributes to the complete assembly of 20S proteasome complex. In particular, proteasome subunit beta type-5, along with other beta subunits, assemble into two heptameric rings and subsequently a proteolytic chamber for substrate degradation. This protein contains "chymotrypsin-like" activity and is capable of cleaving after large hydrophobic residues of peptide. The eukaryotic proteasome recognized degradable proteins, including damaged proteins for protein quality control purpose or key regulatory protein components for dynamic biological processes. An essential function of a modified proteasome, the immunoproteasome, is the processing of class I MHC peptides.

Proteasome subunit alpha type-2 is a protein that in humans is encoded by the PSMA2 gene. This protein is one of the 17 essential subunits that contributes to the complete assembly of 20S proteasome complex.

26S proteasome non-ATPase regulatory subunit 7, also known as 26S proteasome non-ATPase subunit Rpn8, is an enzyme that in humans is encoded by the PSMD7 gene.
Proteasome subunit alpha type-5 also known as 20S proteasome subunit alpha-5 is a protein that in humans is encoded by the PSMA5 gene. This protein is one of the 17 essential subunits that contributes to the complete assembly of 20S proteasome complex.
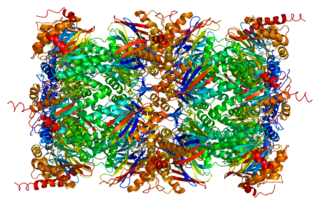
Proteasome subunit beta type-7 as known as 20S proteasome subunit beta-2 is a protein that in humans is encoded by the PSMB7 gene.

26S proteasome non-ATPase regulatory subunit 11 is an enzyme that in humans is encoded by the PSMD11 gene.

Proteasome subunit beta type-6 also known as 20S proteasome subunit beta-1 is a protein that in humans is encoded by the PSMB6 gene.

26S proteasome non-ATPase regulatory subunit 9 is an enzyme that in humans is encoded by the PSMD9 gene.
























