
In biochemistry, a ribonucleotide is a nucleotide containing ribose as its pentose component. It is considered a molecular precursor of nucleic acids. Nucleotides are the basic building blocks of DNA and RNA. Ribonucleotides themselves are basic monomeric building blocks for RNA. Deoxyribonucleotides, formed by reducing ribonucleotides with the enzyme ribonucleotide reductase (RNR), are essential building blocks for DNA. There are several differences between DNA deoxyribonucleotides and RNA ribonucleotides. Successive nucleotides are linked together via phosphodiester bonds.
Biosynthesis, i.e., chemical synthesis occurring in biological contexts, is a term most often referring to multi-step, enzyme-catalyzed processes where chemical substances absorbed as nutrients serve as enzyme substrates, with conversion by the living organism either into simpler or more complex products. Examples of biosynthetic pathways include those for the production of amino acids, lipid membrane components, and nucleotides, but also for the production of all classes of biological macromolecules, and of acetyl-coenzyme A, adenosine triphosphate, nicotinamide adenine dinucleotide and other key intermediate and transactional molecules needed for metabolism. Thus, in biosynthesis, any of an array of compounds, from simple to complex, are converted into other compounds, and so it includes both the catabolism and anabolism of complex molecules. Biosynthetic processes are often represented via charts of metabolic pathways. A particular biosynthetic pathway may be located within a single cellular organelle, while others involve enzymes that are located across an array of cellular organelles and structures.
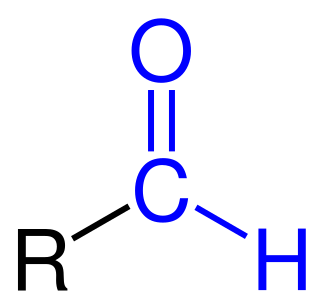
,Formylation refers to any chemical processes in which a compound is functionalized with a formyl group (-CH=O). In organic chemistry, the term is most commonly used with regards to aromatic compounds. In biochemistry the reaction is catalysed by enzymes such as formyltransferases.

Ribose 5-phosphate (R5P) is both a product and an intermediate of the pentose phosphate pathway. The last step of the oxidative reactions in the pentose phosphate pathway is the production of ribulose 5-phosphate. Depending on the body's state, ribulose 5-phosphate can reversibly isomerize to ribose 5-phosphate. Ribulose 5-phosphate can alternatively undergo a series of isomerizations as well as transaldolations and transketolations that result in the production of other pentose phosphates as well as fructose 6-phosphate and glyceraldehyde 3-phosphate.
Phosphoribosylformylglycinamidine cyclo-ligase is the fifth enzyme in the de novo synthesis of purine nucleotides. It catalyzes the reaction to form 5-aminoimidazole ribotide (AIR) from formylglycinamidine-ribonucleotide FGAM. This reaction closes the ring and produces a 5-membered imidazole ring of the purine nucleus (AIR):
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.
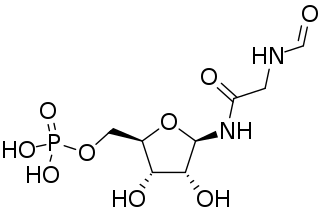
Phosphoribosyl-N-formylglycineamide is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, and hence is a building block for DNA and RNA. The vitamins thiamine and cobalamin also contain fragments derived from FGAR.

10-Formyltetrahydrofolate (10-CHO-THF) is a form of tetrahydrofolate that acts as a donor of formyl groups in anabolism. In these reactions 10-CHO-THF is used as a substrate in formyltransferase reactions.
In enzymology, a formyltetrahydrofolate dehydrogenase (EC 1.5.1.6) is an enzyme that catalyzes the chemical reaction
In enzymology, a 3-methyl-2-oxobutanoate hydroxymethyltransferase (EC 2.1.2.11) is an enzyme that catalyzes the chemical reaction
In enzymology, a glycine formimidoyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a methionyl-tRNA formyltransferase (EC 2.1.2.9) is an enzyme that catalyzes the chemical reaction

In enzymology, a formate—tetrahydrofolate ligase is an enzyme that catalyzes the chemical reaction
In enzymology, an IMP cyclohydrolase (EC 3.5.4.10) is an enzyme that catalyzes the chemical reaction

5′-Phosphoribosyl-5-aminoimidazole is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, and hence is a building block for DNA and RNA. The vitamins thiamine and cobalamin also contain fragments derived from AIR. It is an intermediate in the adenine pathway and is synthesized from 5′-phosphoribosylformylglycinamidine by AIR synthetase.

5-Formamidoimidazole-4-carboxamide ribotide is an intermediate in the formation of purines. It is formed by the enzyme AICAR transformylase from AICAR and 10-formyltetrahydrofolate.

Glycineamide ribonucleotide is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, and hence is a building block for DNA and RNA. The vitamins thiamine and cobalamin also contain fragments derived from GAR.

Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2), also known as glycinamide ribonucleotide transformylase (GAR Tfase), is an enzyme with systematic name 10-formyltetrahydrofolate:5'-phosphoribosylglycinamide N-formyltransferase. This enzyme catalyses the following chemical reaction

The pfl RNA motif refers to a conserved RNA structure present in some bacteria and originally discovered using bioinformatics. pfl RNAs are consistently present in genomic locations that likely correspond to the 5' untranslated regions of protein-coding genes. This arrangement in bacteria is commonly associated with cis-regulatory elements. Moreover, they are in presumed 5' UTRs of multiple non-homologous genes, suggesting that they function only in these locations. Additional evidence of cis-regulatory function came from the observation that predicted rho-independent transcription terminators overlap pfl RNAs. This overlap suggests that the alternate secondary structures of pfl RNA and the transcription terminator stem-loops compete with each other, and this is a common mechanism for cis gene control in bacteria.
UDP-4-amino-4-deoxy-L-arabinose formyltransferase is an enzyme with systematic name 10-formyltetrahydrofolate:UDP-4-amino-4-deoxy-beta-L-arabinose N-formyltransferase. This enzyme catalyses the following chemical reaction













