
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass.
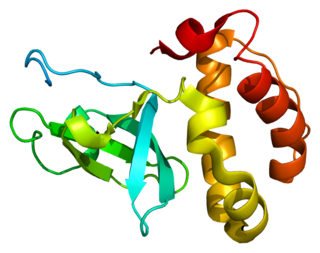
DNA (cytosine-5)-methyltransferase 3 beta, is an enzyme that in humans in encoded by the DNMT3B gene. Mutation in this gene are associated with immunodeficiency, centromere instability and facial anomalies syndrome.
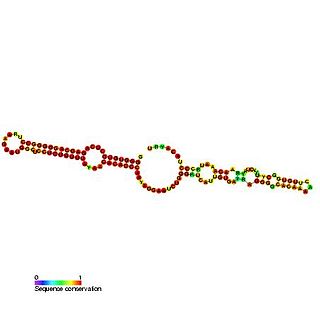
In molecular biology, small nucleolar RNA SNORA11 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA).

In molecular biology, Small nucleolar RNA SNORA77 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA).
In enzymology, a tRNA (guanine-N1-)-methyltransferase (EC 2.1.1.31) is an enzyme that catalyzes the chemical reaction
In enzymology, a tRNA (guanine-N2-)-methyltransferase (EC 2.1.1.32) is an enzyme that catalyzes the chemical reaction

2'-O-methylation is a common nucleoside modification of RNA, where a methyl group is added to the 2' hydroxyl of the ribose moiety of a nucleoside, producing a methoxy group. 2'-O-methylated nucleosides are mostly found in ribosomal RNA and small nuclear RNA and occur in the functionally essential regions of the ribosome and spliceosome. Currently, about 1210 2'-O-methylations (2'-O-Me) have been identified in mammals and yeast and deposited in RMBase database.
23S rRNA (adenine2085-N6)-dimethyltransferase (EC 2.1.1.184, ErmC' methyltransferase, ermC methylase, ermC 23S rRNA methyltransferase, rRNA:m6A methyltransferase ErmC', ErmC', rRNA methyltransferase ErmC' ) is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (adenine2085-N6)-dimethyltransferase. This enzyme catalyses the following chemical reaction
Multisite-specific tRNA:(cytosine-C5)-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (cytosine-C5)-methyltransferase. This enzyme catalyses the following chemical reaction
TRNA (guanine26-N2)-dimethyltransferase (EC 2.1.1.216, Trm1p, TRM1, tRNA (m22G26)dimethyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (guanine26-N2)-dimethyltransferase. This enzyme catalyses the following chemical reaction
TRNA (adenine22-N1)-methyltransferase (EC 2.1.1.217, TrmK, YqfN, Sp1610 (gene), tRNA: m1A22 methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (adenine22-N1)-methyltransferase. This enzyme catalyses the following chemical reaction
TRNA (adenine9-N1)-methyltransferase (EC 2.1.1.218, Trm10p, tRNA(m1G9/m1A9)-methyltransferase, tRNA(m1G9/m1A9)MTase, TK0422p (gene), tRNA m1A9-methyltransferase, tRNA m1A9 Mtase) is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (adenine9-N1)-methyltransferase. This enzyme catalyses the following chemical reaction
TRNA (adenine57-N1/adenine58-N1)-methyltransferase (EC 2.1.1.219, TrmI, PabTrmI, AqTrmI, MtTrmI) is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (adenine57/adenine58-N1)-methyltransferase. This enzyme catalyses the following chemical reaction:
TRNA (adenine58-N1)-methyltransferase (EC 2.1.1.220, tRNA m1A58 methyltransferase, tRNA (m1A58) methyltransferase, TrmI, tRNA (m1A58) Mtase, Rv2118cp, Gcd10p-Gcd14p, Trm61p-Trm6p) is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (adenine58-N1)-methyltransferase. This enzyme catalyses the following chemical reaction
TRNA (guanine9-N1)-methyltransferase (EC 2.1.1.221, Trm10p, tRNA(m1G9/m1A9)-methyltransferase, tRNA(m1G9/m1A9)MTase, tRNA (guanine-N(1)-)-methyltransferase, tRNA m1G9-methyltransferase, tRNA m1G9 MTase) is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (guanine9-N1)-methyltransferase. This enzyme catalyses the following chemical reaction
TRNA (pseudouridine54-N1)-methyltransferase (EC 2.1.1.257, TrmY, m1Psi methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (pseudouridine54-N1)-methyltransferase. This enzyme catalyses the following chemical reaction
Bowen–Conradi syndrome is a disease in humans that can affect children. The disease is due to an autosomal recessive abnormality of the EMG1 gene, which plays a role in small ribosomal subunit (SSU) assembly. The preponderance of diagnoses has been in North American Hutterite children, but BWCNS can affect other population groups.
The Cluster of Excellence Frankfurt "Macromolecular Complexes" (CEF) was established in 2006 by Goethe University Frankfurt together with the Max Planck Institute of Biophysics and the Max Planck Institute for Brain Research in the context of the German Universities Excellence Initiative. Funding by the Deutsche Forschungsgemeinschaft (DFG) endet in October 2019. CEF grew out of the long-standing collaborative research on membrane proteins and RNA molecules and strengthened research efforts in these fields by recruiting further scientists to Frankfurt/Main. CEF brought together the research activities of up to 45 research groups, the majority of which were based on Riedberg Campus in Frankfurt/Main. CEF founded the Buchmann Institute for Molecular Life Sciences (BMLS).
A nucleoside-modified messenger RNA (modRNA) is a synthetic messenger RNA (mRNA) in which some nucleosides are replaced by other naturally modified nucleosides or by synthetic nucleoside analogues. modRNA is used to induce the production of a desired protein in certain cells. An important application is the development of mRNA vaccines, of which the first authorized were COVID-19 vaccines.

N1-Methylpseudouridine is a natural archaeal tRNA component as well as a synthetic pyrimidine nucleoside used in biochemistry and molecular biology for in vitro transcription and is found in the SARS-CoV-2 mRNA vaccines tozinameran (Pfizer–BioNTech) and elasomeran (Moderna).






