Related Research Articles

Electrospray ionization (ESI) is a technique used in mass spectrometry to produce ions using an electrospray in which a high voltage is applied to a liquid to create an aerosol. It is especially useful in producing ions from macromolecules because it overcomes the propensity of these molecules to fragment when ionized. ESI is different from other ionization processes since it may produce multiple-charged ions, effectively extending the mass range of the analyser to accommodate the kDa-MDa orders of magnitude observed in proteins and their associated polypeptide fragments.

In mass spectrometry, matrix-assisted laser desorption/ionization (MALDI) is an ionization technique that uses a laser energy-absorbing matrix to create ions from large molecules with minimal fragmentation. It has been applied to the analysis of biomolecules and various organic molecules, which tend to be fragile and fragment when ionized by more conventional ionization methods. It is similar in character to electrospray ionization (ESI) in that both techniques are relatively soft ways of obtaining ions of large molecules in the gas phase, though MALDI typically produces far fewer multi-charged ions.

Electron-capture dissociation (ECD) is a method of fragmenting gas-phase ions for structure elucidation of peptides and proteins in tandem mass spectrometry. It is one of the most widely used techniques for activation and dissociation of mass selected precursor ion in MS/MS. It involves the direct introduction of low-energy electrons to trapped gas-phase ions.

The history of mass spectrometry has its roots in physical and chemical studies regarding the nature of matter. The study of gas discharges in the mid 19th century led to the discovery of anode and cathode rays, which turned out to be positive ions and electrons. Improved capabilities in the separation of these positive ions enabled the discovery of stable isotopes of the elements. The first such discovery was with the element neon, which was shown by mass spectrometry to have at least two stable isotopes: 20Ne and 22Ne. Mass spectrometers were used in the Manhattan Project for the separation of isotopes of uranium necessary to create the atomic bomb.
Soft laser desorption (SLD) is laser desorption of large molecules that results in ionization without fragmentation. "Soft" in the context of ion formation means forming ions without breaking chemical bonds. "Hard" ionization is the formation of ions with the breaking of bonds and the formation of fragment ions.

MALDI mass spectrometry imaging (MALDI-MSI) is the use of matrix-assisted laser desorption ionization as a mass spectrometry imaging technique in which the sample, often a thin tissue section, is moved in two dimensions while the mass spectrum is recorded. Advantages, like measuring the distribution of a large amount of analytes at one time without destroying the sample, make it a useful method in tissue-based study.

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important method for the accurate mass determination and characterization of proteins, and a variety of methods and instrumentations have been developed for its many uses. Its applications include the identification of proteins and their post-translational modifications, the elucidation of protein complexes, their subunits and functional interactions, as well as the global measurement of proteins in proteomics. It can also be used to localize proteins to the various organelles, and determine the interactions between different proteins as well as with membrane lipids.
Mass spectrometry imaging (MSI) is a technique used in mass spectrometry to visualize the spatial distribution of molecules, as biomarkers, metabolites, peptides or proteins by their molecular masses. After collecting a mass spectrum at one spot, the sample is moved to reach another region, and so on, until the entire sample is scanned. By choosing a peak in the resulting spectra that corresponds to the compound of interest, the MS data is used to map its distribution across the sample. This results in pictures of the spatially resolved distribution of a compound pixel by pixel. Each data set contains a veritable gallery of pictures because any peak in each spectrum can be spatially mapped. Despite the fact that MSI has been generally considered a qualitative method, the signal generated by this technique is proportional to the relative abundance of the analyte. Therefore, quantification is possible, when its challenges are overcome. Although widely used traditional methodologies like radiochemistry and immunohistochemistry achieve the same goal as MSI, they are limited in their abilities to analyze multiple samples at once, and can prove to be lacking if researchers do not have prior knowledge of the samples being studied. Most common ionization technologies in the field of MSI are DESI imaging, MALDI imaging and secondary ion mass spectrometry imaging.
Michael T. Bowers is an American mass spectroscopist, a professor in the Department of Chemistry and Biochemistry at the University of California, Santa Barbara faculty.

Laser spray ionization refers to one of several methods for creating ions using a laser interacting with a spray of neutral particles or ablating material to create a plume of charged particles. The ions thus formed can be separated by m/z with mass spectrometry. Laser spray is one of several ion sources that can be coupled with liquid chromatography-mass spectrometry for the detection of larger molecules.

Matrix-assisted laser desorption electrospray ionization (MALDESI) was first introduced in 2006 as a novel ambient ionization technique which combines the benefits of electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). An infrared (IR) or ultraviolet (UV) laser can be utilized in MALDESI to resonantly excite an endogenous or exogenous matrix. The term ‘matrix’ refers to any molecule that is present in large excess and absorbs the energy of the laser, thus facilitating desorption of analyte molecules. The original MALDESI design was implemented using common organic matrices, similar to those used in MALDI, along with a UV laser. The current MALDESI source employs endogenous water or a thin layer of exogenously deposited ice as the energy-absorbing matrix where O-H symmetric and asymmetric stretching bonds are resonantly excited by a mid-IR laser.

Ambient ionization is a form of ionization in which ions are formed in an ion source outside the mass spectrometer without sample preparation or separation. Ions can be formed by extraction into charged electrospray droplets, thermally desorbed and ionized by chemical ionization, or laser desorbed or ablated and post-ionized before they enter the mass spectrometer.

Catherine Clarke Fenselau is an American scientist who was the first trained mass spectrometrist on the faculty of an American medical school; she joined Johns Hopkins School of Medicine in 1968. She specializes in biomedical applications of mass spectrometry. She has been recognized as an outstanding scientist in the field of bioanalytical chemistry because of her work using mass spectrometry to study biomolecules.
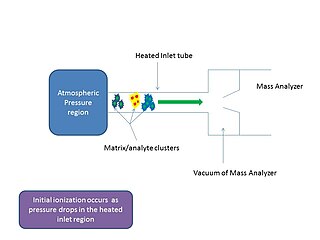
In mass spectrometry, matrix-assisted ionization is a low fragmentation (soft) ionization technique which involves the transfer of particles of the analyte and matrix sample from atmospheric pressure (AP) to the heated inlet tube connecting the AP region to the vacuum of the mass analyzer.
Peter Nemes, Ph.D., is a Hungarian-American chemist, who is active in the fields of bioanalytical chemistry, mass spectrometry, cell/developmental biology, neuroscience, and biochemistry.
The Biemann Medal is awarded annually by the American Society for Mass Spectrometry (ASMS) to an individual early in his or her career in recognition of significant achievement in basic or applied mass spectrometry. It is named after professor Klaus Biemann.

Ron M.A. Heeren is a Dutch scientist in mass spectrometry imaging. He is currently a distinguished professor at Maastricht University and the scientific director of the Multimodal Molecular Imaging Institute (M4I), where he heads the division of Imaging Mass Spectrometry.

Julia Laskin is the William F. and Patty J. Miller Professor of Analytical Chemistry at Purdue University. Her research is focused on the fundamental understanding of ion-surface collisions, understanding of phenomena underlying chemical analysis of large molecules in complex heterogeneous environments, and the development of new instrumentation and methods in preparative and imaging mass spectrometry.
References
- 1 2 3 4 5 Gross, Michael L.; Caprioli, Richard M., eds. (2003). The Encyclopedia of Mass Spectrometry. Elsevier Science. ISBN 978-0-08-043803-0.[ page needed ][ non-primary source needed ]
- ↑ Emmett MR, Caprioli RM (July 1994). "Micro-electrospray mass spectrometry: Ultra-high-sensitivity analysis of peptides and proteins". J Am Soc Mass Spectrom. 5 (7): 605–13. doi: 10.1016/1044-0305(94)85001-1 . PMID 24221962. S2CID 44776275.
- ↑ Caprioli RM, Farmer TB, Gile J (December 1997). "Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS". Anal Chem . 69 (23): 4751–60. doi:10.1021/ac970888i. PMID 9406525.
- ↑ Norris JL, Bierbaum VM (June 2015). "Focus on Imaging Mass Spectrometry, Honoring Dr. Richard M. Caprioli, Recipient of the 2014 ASMS Award for a Distinguished Contribution in Mass Spectrometry". J Am Soc Mass Spectrom. 26 (6): 847–9. Bibcode:2015JASMS..26..847N. doi:10.1007/s13361-015-1138-6. PMID 25893272. S2CID 22023967.