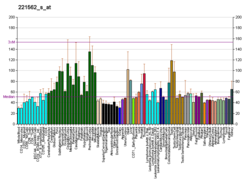
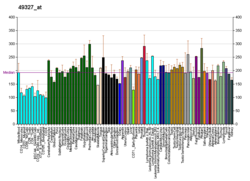
NAD-dependent deacetylase sirtuin-3, mitochondrial also known as SIRT3 is a protein that in humans is encoded by the SIRT3 gene [sirtuin (silent mating type information regulation 2 homolog) 3 (S. cerevisiae)]. [5] [6] SIRT3 is member of the mammalian sirtuin family of proteins, which are homologs to the yeast Sir2 protein. SIRT3 exhibits NAD+-dependent deacetylase activity.
Members of the sirtuin family are characterized by a sirtuin core domain and grouped into four classes, and the protein encoded by this gene is included in class I of the sirtuin family. [5] The human sirtuins have a range of molecular functions and have emerged as important proteins in aging, stress resistance, and metabolic regulation. Yeast sirtuin proteins are known to regulate epigenetic gene silencing and suppress recombination of rDNA. [7] In addition to protein deacetylation, studies have shown that the human sirtuins may also function as intracellular regulatory proteins with mono ADP ribosyltransferase activity.