| Small Cajal body specific RNA 1 | |
|---|---|
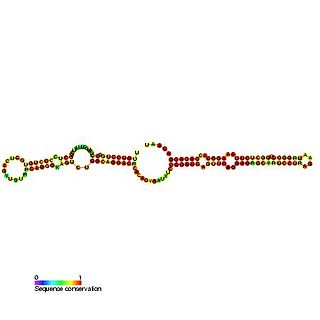
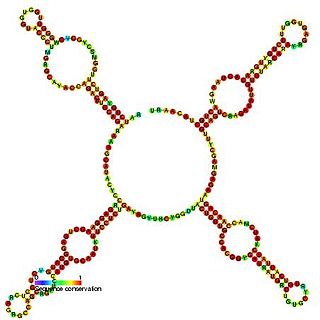
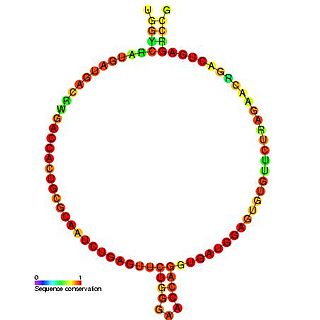
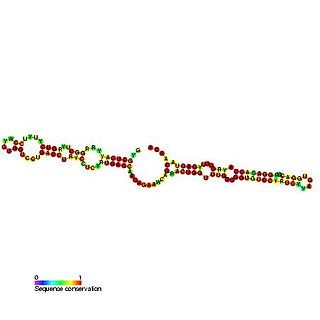
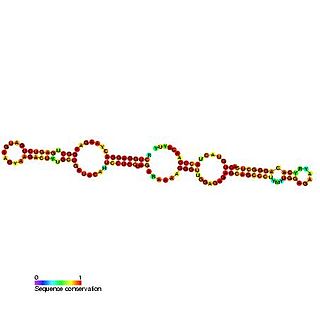
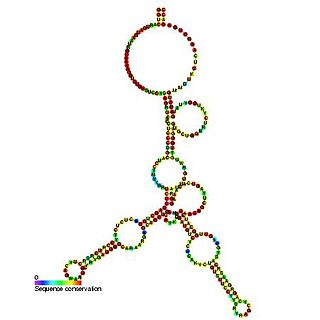
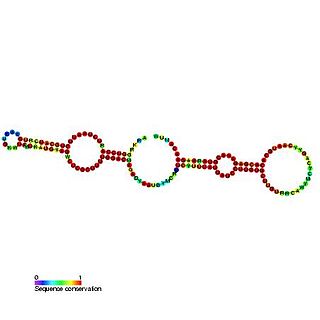
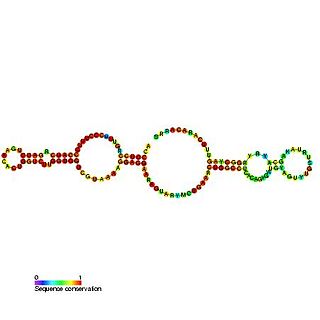
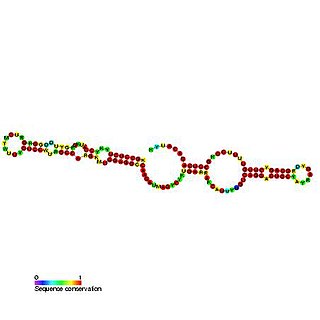
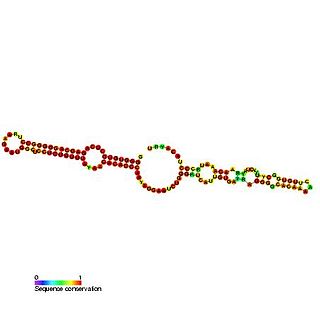
 Predicted secondary structure of Small Cajal body specific RNA 1 | |
| Identifiers | |
| Symbol | SCARNA1 |
| Rfam | RF00553 |
| Other data | |
| RNA type | gene, snRNA, snoRNA, scaRNA; |
| Domain(s) | Vertebrata |
| PDB structures | PDBe |
Small Cajal body-specific RNAs (scaRNAs) are a class of small nucleolar RNAs (snoRNAs) that specifically localise to the Cajal body, a nuclear organelle (cellular sub-organelle) involved in the biogenesis of small nuclear ribonucleoproteins (snRNPs or snurps). ScaRNAs guide the modification (methylation and pseudouridylation) of RNA polymerase II transcribed spliceosomal RNAs U1, U2, U4, U5 and U12.
Small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs, which are associated with methylation, and the H/ACA box snoRNAs, which are associated with pseudouridylation. SnoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes.

Cajal bodies (CBs) also coiled bodies, are spherical nuclear bodies of 0.3–1.0 µm in diameter found in the nucleus of proliferative cells like embryonic cells and tumor cells, or metabolically active cells like neurons. CBs are membrane-less organelles and largely consist of proteins and RNA. They were first reported by Santiago Ramón y Cajal in 1903, who called them nucleolar accessory bodies due to their association with the nucleoli in neuronal cells. They were rediscovered with the use of the electron microscope (EM) and named coiled bodies, according to their appearance as coiled threads on EM images, and later renamed after their discoverer. Research on CBs was accelerated after discovery and cloning of the marker protein p80/Coilin. CBs have been implicated in RNA-related metabolic processes such as the biogenesis, maturation and recycling of snRNPs, histone mRNA processing and telomere maintenance. CBs assemble RNA which is used by telomerase to add nucleotides to the ends of telomeres.

In cell biology, the nucleus is a membrane-bound organelle found in eukaryotic cells. Eukaryotes usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoclasts have many.
Contents
The first scaRNA identified was U85. [1] It is unlike typical snoRNAs in that it is a composite C/D box and H/ACA box snoRNAs and can guide both pseudouridylation and 2'-O-methylation. Not all scaRNAs are composite C/D and H/ACA box snoRNA and most scaRNAs are structurally and functionally indistinguishable from snoRNAs, directing ribosomal RNA (rRNA) modification in the nucleolus.

Ribosomal ribonucleic acid (rRNA) is the RNA component of the ribosome, and is essential for protein synthesis in all living organisms. It constitutes the predominant material within the ribosome, which is approximately 60% rRNA and 40% protein by weight, or 3/5 of ribosome mass. Ribosomes contain two major rRNAs and 50 or more proteins. The ribosomal RNAs form two subunits, the large subunit (LSU) and small subunit (SSU). The LSU rRNA acts as a ribozyme, catalyzing peptide bond formation. The SSU and LSU rRNA sequences are widely used for working out evolutionary relationships among organisms, since they are of ancient origin and are found in all known forms of life.
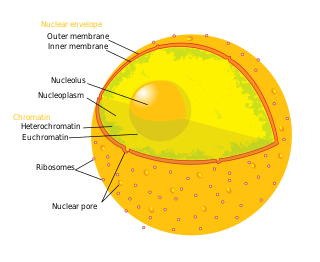
The nucleolus is the largest structure in the nucleus of eukaryotic cells. It is best known as the site of ribosome biogenesis. Nucleoli also participate in the formation of signal recognition particles and play a role in the cell's response to stress. Nucleoli are made of proteins, DNA and RNA and form around specific chromosomal regions called nucleolar organizing regions. Malfunction of nucleoli can be the cause of several human conditions called "nucleolopathies" and the nucleolus is being investigated as a target for cancer chemotherapy.