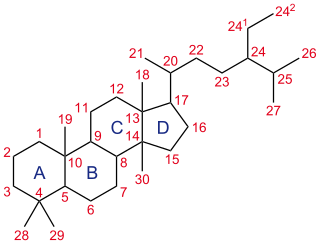
A steroid is a biologically active organic compound with four rings arranged in a specific molecular configuration. Steroids have two principal biological functions: as important components of cell membranes that alter membrane fluidity; and as signaling molecules. Hundreds of steroids are found in plants, animals and fungi. All steroids are manufactured in cells from the sterols lanosterol (opisthokonts) or cycloartenol (plants). Lanosterol and cycloartenol are derived from the cyclization of the triterpene squalene.

Squalene is an organic compound. It is a triterpenoid with the formula C30H50. It is a colourless oil, although impure samples appear yellow. It was originally obtained from shark liver oil (hence its name, as Squalus is a genus of sharks). An estimated 12% of bodily squalene in humans is found in sebum. Squalene has a role in topical skin lubrication and protection.

Hopanoids are a diverse subclass of triterpenoids with the same hydrocarbon skeleton as the compound hopane. This group of pentacyclic molecules therefore refers to simple hopenes, hopanols and hopanes, but also to extensively functionalized derivatives such as bacteriohopanepolyols (BHPs) and hopanoids covalently attached to lipid A.
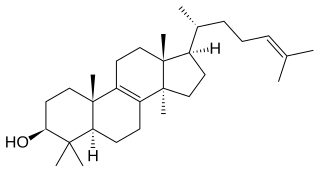
Lanosterol is a tetracyclic triterpenoid and is the compound from which all animal and fungal steroids are derived. By contrast plant steroids are produced via cycloartenol.

Sterane (cyclopentanoperhydrophenanthrenes) compounds are a class of tetracyclic triterpanes derived from steroids or sterols via diagenetic and catagenetic degradation, such as hydrogenation. Steranes are detected in sediments and sedimentary rocks in nature. Steranes have an androstane skeleton with a side chain at carbon C-17. The sterane structure constitutes the core of all sterols. Steranes are widely used as biomarkers for the presence of eukaryotes in past ecosystems because steroids are nearly exclusively produced by eukaryotes. In particular, cholestanes are diagenetic products of cholesterol in animals, while stigmastanes are diagenetic products of stigmasterols in algae and land plants. However, some bacteria are now known to produce sterols and it is inferred that the ultimate origin of sterol biosynthesis is in bacteria. Sterols are produced via protosterols that are direct cyclization compounds of squalene by the catalysis of oxidosqualene cyclase. All known sterols in eukaryotes are enzymatically extensively modified from protosterols, while organisms that only produce protosterols are not known. The oldest record of modified steranes are in sedimentary rocks deposited ca. 720–820 million years ago. In contrast, diagenetic products of protosterols are widely distributed in older Proterozoic rocks and imply the presence of extinct proto-eukaryotes and/or sterol-producing bacteria before the evolution of crown-group eukaryotes.

Triterpenes are a class of terpenes composed of six isoprene units with the molecular formula C30H48; they may also be thought of as consisting of three terpene units. Animals, plants and fungi all produce triterpenes, including squalene, the precursor to all steroids.
Squalene synthase (SQS) or farnesyl-diphosphate:farnesyl-diphosphate farnesyl transferase is an enzyme localized to the membrane of the endoplasmic reticulum. SQS participates in the isoprenoid biosynthetic pathway, catalyzing a two-step reaction in which two identical molecules of farnesyl pyrophosphate (FPP) are converted into squalene, with the consumption of NADPH. Catalysis by SQS is the first committed step in sterol synthesis, since the squalene produced is converted exclusively into various sterols, such as cholesterol, via a complex, multi-step pathway. SQS belongs to squalene/phytoene synthase family of proteins.
(S)-2,3-Oxidosqualene ((S)-2,3-epoxysqualene) is an intermediate in the synthesis of the cell membrane sterol precursors lanosterol and cycloartenol, as well as saponins. It is formed when squalene is oxidized by the enzyme squalene monooxygenase. 2,3-Oxidosqualene is the substrate of various oxidosqualene cyclases, including lanosterol synthase, which produces lanosterol, a precursor to cholesterol.

Lanosterol synthase is an oxidosqualene cyclase (OSC) enzyme that converts (S)-2,3-oxidosqualene to a protosterol cation and finally to lanosterol. Lanosterol is a key four-ringed intermediate in cholesterol biosynthesis. In humans, lanosterol synthase is encoded by the LSS gene.

Isopentenyl pyrophosphate isomerase, also known as Isopentenyl-diphosphate delta isomerase, is an isomerase that catalyzes the conversion of the relatively un-reactive isopentenyl pyrophosphate (IPP) to the more-reactive electrophile dimethylallyl pyrophosphate (DMAPP). This isomerization is a key step in the biosynthesis of isoprenoids through the mevalonate pathway and the MEP pathway.

Prenyltransferases (PTs) are a class of enzymes that transfer allylic prenyl groups to acceptor molecules. Prenyl transferases commonly refer to isoprenyl diphosphate syntheses (IPPSs). Prenyltransferases are a functional category and include several enzyme groups that are evolutionarily independent.
Squalene—hopanol cyclase (EC 4.2.1.129, squalene—hopene cyclase) is an enzyme with systematic name hopan-22-ol hydro-lyase. This enzyme catalyses the following chemical reaction
β-amyrin synthase is an enzyme with systematic name (3S)-2,3-epoxy-2,3-dihydrosqualene mutase . This enzyme catalyses the following chemical reaction
Lupeol synthase is an enzyme with systematic name (3S)-2,3-epoxy-2,3-dihydrosqualene mutase . This enzyme catalyses the following chemical reaction

Parkeol is a relatively uncommon sterol secondary metabolite found mostly in plants, particularly noted in Butyrospermum parkii. It can be synthesized as a minor product by several oxidosqualene cyclase enzymes, and is the sole product of the enzyme parkeol synthase.

Oxidosqualene cyclases (OSC) are enzymes involved in cyclization reactions of 2,3-oxidosqualene to form sterols or triterpenes.

Taraxasterol (anthesterin) is a triterpene derived from the mevalonate pathway and is found in dandelions.

Tetrahymanol is a gammacerane-type membrane lipid first found in the marine ciliate Tetrahymena pyriformis. It was later found in other ciliates, fungi, ferns, and bacteria. After being deposited in sediments that compress into sedimentary rocks over millions of years, tetrahymanol is dehydroxylated into gammacerane. Gammacerane has been interpreted as a proxy for ancient water column stratification.

Isoarborinol is a triterpenoid ubiquitously produced by angiosperms and is thus considered a biomarker for higher plants. Though no isoarborinol-producing microbe has been identified, isoarborinol is also considered a possible biomarker for marine bacteria, as its diagenetic end product, arborane, has been found in ancient marine sediments that predate the rise of plants. Importantly, isoarborinol may represent the phylogenetic link between hopanols and sterols.

Diplopterol is a triterpenoid molecule commonly produced by bacteria, ferns, and a few protozoans. This compound, classified as a member of the hopanoid family, is synthesized from triterpenoid precursor squalene. It is generally believed that hopanoids serve a similar function in bacteria as that of sterols in eukaryotes, which involves modulating membrane fluidity. Diplopterol serves as a useful biomarker for prokaryotic life, along with oxygen content at the time of sediment deposition.