Related Research Articles
Methanogens are anaerobic archaea that produce methane as a byproduct of their energy metabolism, i.e., catabolism. Methane production, or methanogenesis, is the only biochemical pathway for ATP generation in methanogens. All known methanogens belong exclusively to the domain Archaea, although some bacteria, plants, and animal cells are also known to produce methane. However, the biochemical pathway for methane production in these organisms differs from that in methanogens and does not contribute to ATP formation. Methanogens belong to various phyla within the domain Archaea. Previous studies placed all known methanogens into the superphylum Euryarchaeota. However, recent phylogenomic data have led to their reclassification into several different phyla. Methanogens are common in various anoxic environments, such as marine and freshwater sediments, wetlands, the digestive tracts of animals, wastewater treatment plants, rice paddy soil, and landfills. While some methanogens are extremophiles, such as Methanopyrus kandleri, which grows between 84 and 110°C, or Methanonatronarchaeum thermophilum, which grows at a pH range of 8.2 to 10.2 and a Na+ concentration of 3 to 4.8 M, most of the isolates are mesophilic and grow around neutral pH.
Methanogenesis or biomethanation is the formation of methane coupled to energy conservation by microbes known as methanogens. It is the fourth and final stage of anaerobic digestion. Organisms capable of producing methane for energy conservation have been identified only from the domain Archaea, a group phylogenetically distinct from both eukaryotes and bacteria, although many live in close association with anaerobic bacteria. The production of methane is an important and widespread form of microbial metabolism. In anoxic environments, it is the final step in the decomposition of biomass. Methanogenesis is responsible for significant amounts of natural gas accumulations, the remainder being thermogenic.

Sulfate-reducing microorganisms (SRM) or sulfate-reducing prokaryotes (SRP) are a group composed of sulfate-reducing bacteria (SRB) and sulfate-reducing archaea (SRA), both of which can perform anaerobic respiration utilizing sulfate (SO2−
4) as terminal electron acceptor, reducing it to hydrogen sulfide (H2S). Therefore, these sulfidogenic microorganisms "breathe" sulfate rather than molecular oxygen (O2), which is the terminal electron acceptor reduced to water (H2O) in aerobic respiration.
An acetogen is a microorganism that generates acetate (CH3COO−) as an end product of anaerobic respiration or fermentation. However, this term is usually employed in a narrower sense only to those bacteria and archaea that perform anaerobic respiration and carbon fixation simultaneously through the reductive acetyl coenzyme A (acetyl-CoA) pathway (also known as the Wood-Ljungdahl pathway). These genuine acetogens are also known as "homoacetogens" and they can produce acetyl-CoA (and from that, in most cases, acetate as the end product) from two molecules of carbon dioxide (CO2) and four molecules of molecular hydrogen (H2). This process is known as acetogenesis, and is different from acetate fermentation, although both occur in the absence of molecular oxygen (O2) and produce acetate. Although previously thought that only bacteria are acetogens, some archaea can be considered to be acetogens.
Acetogenesis is a process through which acetyl-CoA or acetic acid is produced by anaerobic bacteria through the reduction of CO2 via the Wood–Ljungdahl pathway. Other microbial processes that produce acetic acid are not considered acetogenesis. The diverse bacterial species capable of acetogenesis are collectively called acetogens.
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.

Methanobacterium is a genus of the Methanobacteria class in the Archaea kingdom, which produce methane as a metabolic byproduct. Despite the name, this genus belongs not to the bacterial domain but the archaeal domain. Methanobacterium are nonmotile and live without oxygen, which is toxic to them, and they only inhabit anoxic environments.
Anaerobic oxidation of methane (AOM) is a methane-consuming microbial process occurring in anoxic marine and freshwater sediments. AOM is known to occur among mesophiles, but also in psychrophiles, thermophiles, halophiles, acidophiles, and alkophiles. During AOM, methane is oxidized with different terminal electron acceptors such as sulfate, nitrate, nitrite and metals, either alone or in syntrophy with a partner organism.
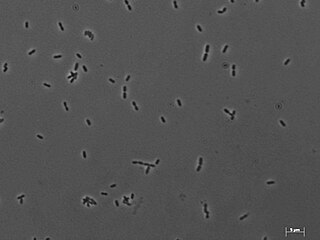
Methanobrevibacter smithii is the predominant methanogenic archaeon in the microbiota of the human gut. M. smithii has a coccobacillus shape. It plays an important role in the efficient digestion of polysaccharides (complex sugars) by consuming the end products of bacterial fermentation (H2, CO2, acetate, and formate). M. smithii is a hydrogenotrophic methanogen that utilizes hydrogen by combining it with carbon dioxide to form methane. The removal of hydrogen by M. smithii is thought to allow an increase in the extraction of energy from nutrients by shifting bacterial fermentation to more oxidized end products.
In the taxonomy of microorganisms, the Methanothrix is a genus of methanogenic archaea within the Euryarchaeota. Methanothrix cells were first isolated from a mesophilic sewage digester but have since been found in many anaerobic and aerobic environments. Methanothrix were originally understood to be obligate anaerobes that can survive exposure to high concentrations of oxygen, but recent studies have shown at least one Candidatus operational taxonomic unit proposed to be in the Methanothrix genus not only survives but remains active in oxic soils. This proposed species, Ca. Methanothrix paradoxum, is frequently found in methane-releasing ecosystems and is the dominant methanogen in oxic soils.
Syntrophobacter wolinii is a non-motile, gram-negative and rod-shaped species of bacteria that was originally isolated from a wastewater digester. This species is able to perform propionate degradation and sulfate reduction. S. wolinii can be grown in co-culture or pure culture. 16s rRNA analysis shows its close relation to other sulfate reducers.

Methanococcus maripaludis is a species of methanogenic archaea found in marine environments, predominantly salt marshes. M. maripaludis is a non-pathogenic, gram-negative, weakly motile, non-spore-forming, and strictly anaerobic mesophile. It is classified as a chemolithoautotroph. This archaeon has a pleomorphic coccoid-rod shape of 1.2 by 1.6 μm, in average size, and has many unique metabolic processes that aid in survival. M. maripaludis also has a sequenced genome consisting of around 1.7 Mbp with over 1,700 identified protein-coding genes. In ideal conditions, M. maripaludis grows quickly and can double every two hours.
Syntrophomonas zehnderi is a bacterium. It is anaerobic, syntrophic and fatty acid-oxidizing. The type strain is OL-4T. Cells are slightly curved, non-motile rods.
Syntrophus aciditrophicus is a gram-negative and rod-shaped bacterium. It is non-motile, non-spore-forming and grows under strictly anaerobic conditions, thus an obligate anaerobe. It degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Its genome was published in 2007.
Syntrophococcus sucromutans is a Gram-negative strictly anaerobic chemoorganotrophic Bacillota. These bacteria can be found forming small chains in the habitat where it was first isolated, the rumen of cows. It is the type strain of genus Syntrophococcus and it has an uncommon one-carbon metabolic pathway, forming acetate from formate as a product of sugar oxidation.
Interspecies hydrogen transfer (IHT) is a form of interspecies electron transfer. It is a syntrophic process by which H2 is transferred from one organism to another, particularly in the rumen and other anaerobic environments.
Desulfovibrio alcoholivorans is a bacterium from the genus of Desulfovibrio which has been isolated from alcohol industry waste water in France.
Biological methanation (also: biological hydrogen methanation (BHM) or microbiological methanation) is a conversion process to generate methane by means of highly specialized microorganisms (Archaea) within a technical system. This process can be applied in a power-to-gas system to produce biomethane and is appreciated as an important storage technology for variable renewable energy in the context of energy transition. This technology was successfully implemented at a first power-to-gas plant of that kind in the year 2015.
Formatotrophs are organisms that can assimilate formate or formic acid to use as a carbon source or for reducing power. Some authors classify formatotrophs as one of the five trophic groups of methanogens, which also include hydrogenotrophs, acetotrophs, methylotrophs, and alcoholotrophs. Formatotrophs have garnered attention for applications in biotechnology as part of a "formate bioeconomy" in which synthesized formate could be used as a nutrient for microoganisms. Formate can be electrochemically synthesized from CO2 and renewable energy, and formatotrophs may be genetically modified to enhance production of biochemical products to be used as biofuels. Technical limitations in culturing formatotrophs have limited the discovery of natural formatotrophs and impeded research on their formate-metabolizing enzymes, which are of interest for applications in carbon sequestration and astrobiology.
Syntrophaceae is a family of bacteria in the order Syntrophales. It comprises four genera: Syntrophus, Smithella, Desulfobacca and Desulfomonile. All members have rod-shaped cells and do not form spores. As gram-negative bacteria, the presence of a thin cell wall and a double membrane is notable. This bacterium usually forms red or pink colonies.
References
- 1 2 3 Gentry, Terry J.; Pepper, Ian L.; Pierson, Leland S. (2015-01-01), Pepper, Ian L.; Gerba, Charles P.; Gentry, Terry J. (eds.), "Chapter 19 - Microbial Diversity and Interactions in Natural Ecosystems", Environmental Microbiology (Third Edition), San Diego: Academic Press, pp. 441–460, doi:10.1016/b978-0-12-394626-3.00019-3, ISBN 978-0-12-394626-3 , retrieved 2023-12-27
- 1 2 Marietou, Angeliki (2021-01-01), Gadd, Geoffrey Michael; Sariaslani, Sima (eds.), "Chapter Two - Sulfate reducing microorganisms in high temperature oil reservoirs", Advances in Applied Microbiology, 116, Academic Press: 99–131, doi:10.1016/bs.aambs.2021.03.004, PMID 34353505 , retrieved 2023-12-27
- 1 2 3 4 5 Schink B, Stams AJ (2013). "Syntrophism Among Prokaryotes". In Rosenberg E, DeLong EF, Lory S, Stackebrandt E (eds.). The Prokaryotes: Prokaryotic Communities and Ecophysiology. Berlin, Heidelberg: Springer. pp. 471–493. doi:10.1007/978-3-642-30123-0_59. ISBN 978-3-642-30123-0.
- 1 2 3 Kamagata Y (2015-03-15). "Syntrophy in Anaerobic Digestion". Anaerobic Biotechnology. Imperial College Press. pp. 13–30. doi:10.1142/9781783267910_0002. ISBN 978-1-78326-790-3 . Retrieved 2022-11-11.
- 1 2 "syntrophism | biology | Britannica". 2022-09-30. Archived from the original on 2022-09-30. Retrieved 2023-12-27.
- ↑ "Syntrophism Definition & Meaning | Merriam-Webster Medical". 2022-08-19. Archived from the original on 2022-08-19. Retrieved 2023-12-27.
- ↑ Hao L, Michaelsen TY, Singleton CM, Dottorini G, Kirkegaard RH, Albertsen M, et al. (April 2020). "Novel syntrophic bacteria in full-scale anaerobic digesters revealed by genome-centric metatranscriptomics". The ISME Journal. 14 (4): 906–918. Bibcode:2020ISMEJ..14..906H. doi:10.1038/s41396-019-0571-0. PMC 7082340 . PMID 31896784.
- ↑ Dolfing J (January 2014). "Syntrophy in microbial fuel cells". The ISME Journal. 8 (1): 4–5. Bibcode:2014ISMEJ...8....4D. doi:10.1038/ismej.2013.198. PMC 3869025 . PMID 24173460.
- 1 2 3 4 5 Morris BE, Henneberger R, Huber H, Moissl-Eichinger C (May 2013). "Microbial syntrophy: interaction for the common good". FEMS Microbiology Reviews. 37 (3): 384–406. doi: 10.1111/1574-6976.12019 . PMID 23480449.
- ↑ Sieber JR, McInerney MJ, Gunsalus RP (2012). "Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation". Annual Review of Microbiology. 66: 429–452. doi:10.1146/annurev-micro-090110-102844. PMID 22803797.
- ↑ McInerney MJ, Sieber JR, Gunsalus RP (December 2009). "Syntrophy in anaerobic global carbon cycles". Current Opinion in Biotechnology. 20 (6): 623–632. doi:10.1016/j.copbio.2009.10.001. PMC 2790021 . PMID 19897353.
- ↑ McInerney MJ, Rohlin L, Mouttaki H, Kim U, Krupp RS, Rios-Hernandez L, et al. (May 2007). "The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth". Proceedings of the National Academy of Sciences of the United States of America. 104 (18): 7600–7605. Bibcode:2007PNAS..104.7600M. doi: 10.1073/pnas.0610456104 . PMC 1863511 . PMID 17442750.
- 1 2 McInerney MJ, Sieber JR, Gunsalus RP (December 2009). "Syntrophy in anaerobic global carbon cycles". Current Opinion in Biotechnology. Chemical biotechnology ● Pharmaceutical biotechnology. 20 (6): 623–632. doi:10.1016/j.copbio.2009.10.001. PMC 2790021 . PMID 19897353.
- 1 2 3 4 5 6 Worm P, Müller N, Plugge CM, Stams AJ, Schink B (2010). "Syntrophy in methanogenic degradation.". (Endo)symbiotic Methanogenic Archaea. Microbiology Monographs. Vol. 19. Berlin, Heidelberg: Springer. pp. 143–173. doi:10.1007/978-3-642-13615-3_9. ISBN 978-3-642-13614-6.
- 1 2 3 4 Jackson BE, McInerney MJ (January 2002). "Anaerobic microbial metabolism can proceed close to thermodynamic limits". Nature. 415 (6870): 454–456. Bibcode:2002Natur.415..454J. doi:10.1038/415454a. PMID 11807560. S2CID 9126984.
- 1 2 3 4 Zhang M, Zang L (2019). "A review of interspecies electron transfer in anaerobic digestion". IOP Conf. Ser: Earth Environ. 310 (4): 042026. Bibcode:2019E&ES..310d2026Z. doi: 10.1088/1755-1315/310/4/042026 . S2CID 202886264.
- ↑ Rotaru AE, Shrestha PM, Liu F, Ueki T, Nevin K, Summers ZM, Lovley DR (November 2012). "Interspecies electron transfer via hydrogen and formate rather than direct electrical connections in cocultures of Pelobacter carbinolicus and Geobacter sulfurreducens". Applied and Environmental Microbiology. 78 (21): 7645–7651. Bibcode:2012ApEnM..78.7645R. doi:10.1128/AEM.01946-12. PMC 3485699 . PMID 22923399.
- 1 2 3 4 Zhang Y, Li C, Yuan Z, Wang R, Angelidaki I, Zhu G (2023-01-15). "Syntrophy mechanism, microbial population, and process optimization for volatile fatty acids metabolism in anaerobic digestion". Chemical Engineering Journal. 452: 139137. Bibcode:2023ChEnJ.45239137Z. doi:10.1016/j.cej.2022.139137. ISSN 1385-8947. S2CID 252205776.
- ↑ Wrede C, Dreier A, Kokoschka S, Hoppert M (2012). "Archaea in symbioses". Archaea. 2012: 596846. doi: 10.1155/2012/596846 . PMC 3544247 . PMID 23326206.
- ↑ Dubé CD, Guiot SR (2015). "Direct Interspecies Electron Transfer in Anaerobic Digestion: A Review". Biogas Science and Technology. Advances in Biochemical Engineering/Biotechnology. Vol. 151. pp. 101–15. doi:10.1007/978-3-319-21993-6_4. ISBN 978-3-319-21992-9. PMID 26337845.
- ↑ "What's a Rumen". AnimalSmart.org. Retrieved 2022-11-21.
- 1 2 Ng F, Kittelmann S, Patchett ML, Attwood GT, Janssen PH, Rakonjac J, Gagic D (September 2016). "An adhesin from hydrogen-utilizing rumen methanogen Methanobrevibacter ruminantium M1 binds a broad range of hydrogen-producing microorganisms". Environmental Microbiology. 18 (9): 3010–3021. doi: 10.1111/1462-2920.13155 . PMID 26643468.
- ↑ Sapkota A (2022-07-12). "Syntrophism or Syntrophy Interaction- Definition, Examples". The Biology Notes. Retrieved 2022-11-21.
- ↑ Kang D, Saha S, Kurade MB, Basak B, Ha G, Jeon B, et al. (July 2021). "Dual-stage pulse-feed operation enhanced methanation of lipidic waste during co-digestion using acclimatized consortia". Renewable and Sustainable Energy Reviews. 145: 111096. doi:10.1016/j.rser.2021.111096. ISSN 1364-0321. S2CID 234830362.
- ↑ Stams AJ, de Bok FA, Plugge CM, van Eekert MH, Dolfing J, Schraa G (March 2006). "Exocellular electron transfer in anaerobic microbial communities". Environmental Microbiology. 8 (3): 371–382. Bibcode:2006EnvMi...8..371S. doi:10.1111/j.1462-2920.2006.00989.x. PMID 16478444.
- 1 2 3 Callaghan AV, Morris BE, Pereira IA, McInerney MJ, Austin RN, Groves JT, et al. (January 2012). "The genome sequence of Desulfatibacillum alkenivorans AK-01: a blueprint for anaerobic alkane oxidation". Environmental Microbiology. 14 (1): 101–113. Bibcode:2012EnvMi..14..101C. doi:10.1111/j.1462-2920.2011.02516.x. PMID 21651686.
- 1 2 3 4 Ferry JG, Wolfe RS (February 1976). "Anaerobic degradation of benzoate to methane by a microbial consortium". Archives of Microbiology. 107 (1): 33–40. Bibcode:1976ArMic.107...33F. doi:10.1007/BF00427864. PMID 1252087. S2CID 31426072.
- 1 2 3 4 Zindel U, Freudenberg W, Rieth M, Andreesen JR, Schnell J, Widdel F (July 1988). "Eubacterium acidaminophilum sp. nov., a versatile amino acid-degrading anaerobe producing or utilizing H2 or formate". Archives of Microbiology. 150 (3): 254–266. Bibcode:1988ArMic.150..254Z. doi:10.1007/BF00407789. ISSN 0302-8933. S2CID 34824309.
- ↑ McInerney MJ, Bryant MP, Hespell RB, Costerton JW (April 1981). "Syntrophomonas wolfei gen. nov. sp. nov., an Anaerobic, Syntrophic, Fatty Acid-Oxidizing Bacterium". Applied and Environmental Microbiology. 41 (4): 1029–1039. Bibcode:1981ApEnM..41.1029M. doi:10.1128/aem.41.4.1029-1039.1981. PMC 243852 . PMID 16345745.
- ↑ Schöcke L, Schink B (September 1998). "Membrane-bound proton-translocating pyrophosphatase of Syntrophus gentianae, a syntrophically benzoate-degrading fermenting bacterium". European Journal of Biochemistry. 256 (3): 589–594. doi:10.1046/j.1432-1327.1998.2560589.x. PMID 9780235.