A dehydrogenase is an enzyme belonging to the group of oxidoreductases that oxidizes a substrate by reducing an electron acceptor, usually NAD+/NADP+ or a flavin coenzyme such as FAD or FMN. Like all catalysts, they catalyze reverse as well as forward reactions, and in some cases this has physiological significance: for example, alcohol dehydrogenase catalyzes the oxidation of ethanol to acetaldehyde in animals, but in yeast it catalyzes the production of ethanol from acetaldehyde.
Acetaldehyde is an organic chemical compound with the formula CH3 CHO, sometimes abbreviated as MeCHO. It is a colorless liquid or gas, boiling near room temperature. It is one of the most important aldehydes, occurring widely in nature and being produced on a large scale in industry. Acetaldehyde occurs naturally in coffee, bread, and ripe fruit, and is produced by plants. It is also produced by the partial oxidation of ethanol by the liver enzyme alcohol dehydrogenase and is a contributing cause of hangover after alcohol consumption. Pathways of exposure include air, water, land, or groundwater, as well as drink and smoke. Consumption of disulfiram inhibits acetaldehyde dehydrogenase, the enzyme responsible for the metabolism of acetaldehyde, thereby causing it to build up in the body.
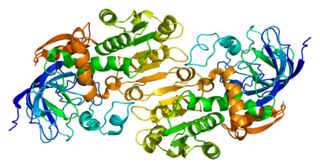
Alcohol dehydrogenases (ADH) (EC 1.1.1.1) are a group of dehydrogenase enzymes that occur in many organisms and facilitate the interconversion between alcohols and aldehydes or ketones with the reduction of nicotinamide adenine dinucleotide (NAD+) to NADH. In humans and many other animals, they serve to break down alcohols that are otherwise toxic, and they also participate in the generation of useful aldehyde, ketone, or alcohol groups during the biosynthesis of various metabolites. In yeast, plants, and many bacteria, some alcohol dehydrogenases catalyze the opposite reaction as part of fermentation to ensure a constant supply of NAD+.

Disulfiram is a medication used to support the treatment of chronic alcoholism by producing an acute sensitivity to ethanol. Disulfiram works by inhibiting the enzyme aldehyde dehydrogenase, causing many of the effects of a hangover to be felt immediately following alcohol consumption. Disulfiram plus alcohol, even small amounts, produces flushing, throbbing in the head and neck, a throbbing headache, respiratory difficulty, nausea, copious vomiting, sweating, thirst, chest pain, palpitation, dyspnea, hyperventilation, fast heart rate, low blood pressure, fainting, marked uneasiness, weakness, vertigo, blurred vision, and confusion. In severe reactions there may be respiratory depression, cardiovascular collapse, abnormal heart rhythms, heart attack, acute congestive heart failure, unconsciousness, convulsions, and death.

Malate dehydrogenase (EC 1.1.1.37) (MDH) is an enzyme that reversibly catalyzes the oxidation of malate to oxaloacetate using the reduction of NAD+ to NADH. This reaction is part of many metabolic pathways, including the citric acid cycle. Other malate dehydrogenases, which have other EC numbers and catalyze other reactions oxidizing malate, have qualified names like malate dehydrogenase (NADP+).

Alcohol flush reaction is a condition in which a person develops flushes or blotches associated with erythema on the face, neck, shoulders, ears, and in some cases, the entire body after consuming alcoholic beverages. The reaction is the result of an accumulation of acetaldehyde, a metabolic byproduct of the catabolic metabolism of alcohol, and is caused by an aldehyde dehydrogenase 2 deficiency.

Alcohol tolerance refers to the bodily responses to the functional effects of ethanol in alcoholic beverages. This includes direct tolerance, speed of recovery from insobriety and resistance to the development of alcohol use disorder.
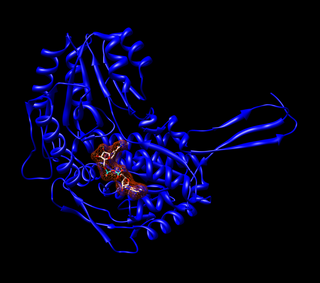
Aldehyde dehydrogenases are a group of enzymes that catalyse the oxidation of aldehydes. They convert aldehydes to carboxylic acids. The oxygen comes from a water molecule. To date, nineteen ALDH genes have been identified within the human genome. These genes participate in a wide variety of biological processes including the detoxification of exogenously and endogenously generated aldehydes.

Aldehyde dehydrogenase, mitochondrial is an enzyme that in humans is encoded by the ALDH2 gene located on chromosome 12. ALDH2 belongs to the aldehyde dehydrogenase family of enzymes. Aldehyde dehydrogenase is the second enzyme of the major oxidative pathway of alcohol metabolism. ALDH2 has a low Km for acetaldehyde, and is localized in mitochondrial matrix. The other liver isozyme, ALDH1, localizes to the cytosol.

In enzymology, a retinal dehydrogenase, also known as retinaldehyde dehydrogenase (RALDH), catalyzes the chemical reaction converting retinal to retinoic acid. This enzyme belongs to the family of oxidoreductases, specifically the class acting on aldehyde or oxo- donor groups with NAD+ or NADP+ as acceptor groups, the systematic name being retinal:NAD+ oxidoreductase. This enzyme participates in retinol metabolism. The general scheme for the reaction catalyzed by this enzyme is:

Alcohol dehydrogenase 1B is an enzyme that in humans is encoded by the ADH1B gene.

Alcohol dehydrogenase 1C is an enzyme that in humans is encoded by the ADH1C gene.

The short-term effects of alcohol consumption range from a decrease in anxiety and motor skills and euphoria at lower doses to intoxication (drunkenness), to stupor, unconsciousness, anterograde amnesia, and central nervous system depression at higher doses. Cell membranes are highly permeable to alcohol, so once it is in the bloodstream, it can diffuse into nearly every cell in the body.

Coprine is a mycotoxin. It was first isolated from common inkcap. It occurs in mushrooms in the genera Coprinopsis. When combined with alcohol, it causes "Coprinus syndrome". It inhibits the enzyme aldehyde dehydrogenase, which is involved in the metabolism of alcohol. This inhibition leads to a buildup of acetaldehyde, causing an alcohol flush reaction. Because of this, the mushroom is commonly referred to as Tippler's Bane.
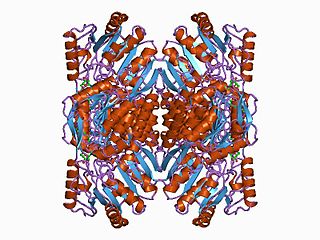
Aldehyde dehydrogenase 1 family, member A1, also known as ALDH1A1 or retinaldehyde dehydrogenase 1 (RALDH1), is an enzyme that is encoded by the ALDH1A1 gene.
Pseudohypoxia refers to a condition that mimics hypoxia, by having sufficient oxygen yet impaired mitochondrial respiration due to a deficiency of necessary co-enzymes, such as NAD+ and TPP. The increased cytosolic ratio of free NADH/NAD+ in cells (more NADH than NAD+) can be caused by diabetic hyperglycemia and by excessive alcohol consumption. Low levels of TPP results from thiamine deficiency.

Alcohol intolerance is due to a genetic polymorphism of the aldehyde dehydrogenase enzyme, which is responsible for the metabolism of acetaldehyde. This polymorphism is most often reported in patients of East Asian descent. Alcohol intolerance may also be an associated side effect of certain drugs such as disulfiram, metronidazole, or nilutamide. Skin flushing and nasal congestion are the most common symptoms of intolerance after alcohol ingestion. It may also be characterized as intolerance causing hangover symptoms similar to the "disulfiram-like reaction" of aldehyde dehydrogenase deficiency or chronic fatigue syndrome. Severe pain after drinking alcohol may indicate a more serious underlying condition.

A disulfiram-like drug is a drug that causes an adverse reaction to alcohol leading to nausea, vomiting, flushing, dizziness, throbbing headache, chest and abdominal discomfort, and general hangover-like symptoms among others. These effects are caused by accumulation of acetaldehyde, a major but toxic metabolite of alcohol formed by the enzyme alcohol dehydrogenase. The reaction has been variously termed a disulfiram-like reaction, alcohol intolerance, and acetaldehyde syndrome.

Alda-1 is an organic compound that enhances the enzymatic activity of human ALDH2. Alda-1 has been proposed as a potential treatment for the alcohol flush reaction experienced by people with genetically deficient ALDH2.

Disulfiram-alcohol reaction (DAR) is the effect of the interaction in the human body of alcohol drunk with disulfiram or some mushrooms. The DAR is key to disulfiram therapy that is widely used for alcohol-aversive treatment and management of other addictions. Once disulfiram-treated patients take alcohol, even in small doses, they experience strong unpleasant sensations.
















