Related Research Articles

Cellulase is any of several enzymes produced chiefly by fungi, bacteria, and protozoans that catalyze cellulolysis, the decomposition of cellulose and of some related polysaccharides:

β-Glucosidase is an enzyme that catalyses the following reaction:

A wood-decay or xylophagous fungus is any species of fungus that digests moist wood, causing it to rot. Some species of wood-decay fungi attack dead wood, such as brown rot, and some, such as Armillaria, are parasitic and colonize living trees. Excessive moisture above the fibre saturation point in wood is required for fungal colonization and proliferation. In nature, this process causes the breakdown of complex molecules and leads to the return of nutrients to the soil. Wood-decay fungi consume wood in various ways; for example, some attack the carbohydrates in wood, and some others decay lignin. The rate of decay of wooden materials in various climates can be estimated by empirical models.
Cellobiohydrolase may refer to:
In enzymology, a beta-galactoside alpha-2,6-sialyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a cellulose synthase (GDP-forming) is an enzyme that catalyzes the chemical reaction
Cels may refer to:

In molecular biology, a carbohydrate-binding module (CBM) is a protein domain found in carbohydrate-active enzymes. The majority of these domains have carbohydrate-binding activity. Some of these domains are found on cellulosomal scaffoldin proteins. CBMs were previously known as cellulose-binding domains. CBMs are classified into numerous families, based on amino acid sequence similarity. There are currently 64 families of CBM in the CAZy database.

In molecular biology, Glycoside hydrolase family 10 is a family of glycoside hydrolases.

In molecular biology, glycoside hydrolase family 48 is a family of glycoside hydrolases.

In molecular biology, glycoside hydrolase family 7 is a family of glycoside hydrolases EC 3.2.1., which are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycoside hydrolases, based on sequence similarity, has led to the definition of >100 different families. This classification is available on the CAZy web site, and also discussed at CAZypedia, an online encyclopedia of carbohydrate active enzymes.
In molecular biology, glycoside hydrolase family 9 is a family of glycoside hydrolases.
In molecular biology, glycoside hydrolase family 6 is a family of glycoside hydrolases EC 3.2.1., which are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycoside hydrolases, based on sequence similarity, has led to the definition of >100 different families. This classification is available on the CAZy web site, and also discussed at CAZypedia, an online encyclopedia of carbohydrate active enzymes.

Glucanases are enzymes that break down large polysaccharides via hydrolysis. The product of the hydrolysis reaction is called a glucan, a linear polysaccharide made of up to 1200 glucose monomers, held together with glycosidic bonds. Glucans are abundant in the endosperm cell walls of cereals such as barley, rye, sorghum, rice, and wheat. Glucanases are also referred to as lichenases, hydrolases, glycosidases, glycosyl hydrolases, and/or laminarinases. Many types of glucanases share similar amino acid sequences but vastly different substrates. Of the known endo-glucanases, 1,3-1,4-β-glucanase is considered the most active.
Lichenase is an enzyme with systematic name (1→3)-(1→4)-β-D-glucan 4-glucanohydrolase. It was named after its activity in on lichenin.
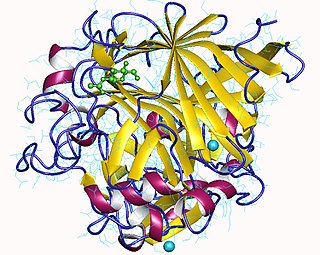
Cellulose 1,4-β-cellobiosidase is an enzyme of interest for its capability of converting cellulose to useful chemicals, particularly cellulosic ethanol.
Oligoxyloglucan reducing-end-specific cellobiohydrolase is an enzyme with systematic name oligoxyloglucan reducing-end cellobiohydrolase. This enzyme catalyses the following chemical reaction
The enzyme exo-(1→4)-α-D-glucan lyase (EC 4.2.2.13, α-(1→4)-glucan 1,5-anhydro-D-fructose eliminase, α-1,4-glucan exo-lyase, α-1,4-glucan lyase, GLase) is an enzyme with systematic name (1→4)-α-D-glucan exo-4-lyase (1,5-anhydro-D-fructose-forming). This enzyme catalyses the following chemical reaction
Mycofactocin (MFT) is a family of small molecules derived from a peptide of the type known as RiPP (ribosomally synthesized and post-translationally modified peptides), naturally occurring in many types of Mycobacterium. It was discovered in a bioinformatics study in 2011. All mycofactocins share a precursor in the form of premycofactocin (PMFT); they differ by the cellulose tail added. Being redox active, both PMFT and MFT have an oxidized dione (mycofactocinone) form and a reduced diol (mycofactocinol) form, respectively termed PMFTH2 and MFTH2.
Cytophaga hutchinsonii is a bacterial species in the genus Cytophaga. C. hutchinsonii is an aerobic, gram-negative, soil, microorganism that exhibits gliding motility, enabling it to move quickly over surfaces and is capable of cellulose degradation.
References
- ↑ Barr BK, Hsieh YL, Ganem B, Wilson DB (January 1996). "Identification of two functionally different classes of exocellulases". Biochemistry. 35 (2): 586–92. doi:10.1021/bi9520388. PMID 8555231.
- ↑ Saharay M, Guo H, Smith JC (October 2010). "Catalytic mechanism of cellulose degradation by a cellobiohydrolase, CelS". PLOS ONE. 5 (10): e12947. doi: 10.1371/journal.pone.0012947 . PMC 2953488 . PMID 20967294.