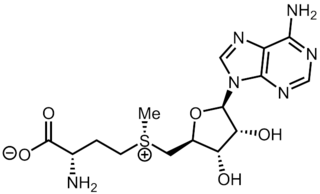
S-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM is produced and consumed in the liver. More than 40 methyl transfers from SAM are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM was first discovered by Giulio Cantoni in 1952.

Spermidine synthase is an enzyme that catalyzes the transfer of the propylamine group from S-adenosylmethioninamine to putrescine in the biosynthesis of spermidine. The systematic name is S-adenosyl 3-(methylthio)propylamine:putrescine 3-aminopropyltransferase and it belongs to the group of aminopropyl transferases. It does not need any cofactors. Most spermidine synthases exist in solution as dimers.
In enzymology, a glycine N-methyltransferase is an enzyme that catalyzes the chemical reaction

Desosamine is a 3-(dimethylamino)-3,4,6-trideoxyhexose found in certain macrolide antibiotics such as the commonly prescribed erythromycin, azithromycin, clarithroymcin, methymycin, narbomycin, oleandomycin, picromycin and roxithromycin. As the name suggests, these macrolide antibiotics contain a macrolide or lactone ring and they are attached to the ring Desosamine which is crucial for bactericidal activity. The biological action of the desosamine-based macrolide antibiotics is to inhibit the bacterial ribosomal protein synthesis. These antibiotics which contain Desosamine are widely used to cure bacterial-causing infections in human respiratory system, skin, muscle tissues, and urethra.

Cobalamin biosynthesis is the process by which bacteria and archea make cobalamin, vitamin B12. Many steps are involved in converting aminolevulinic acid via uroporphyrinogen III and adenosylcobyric acid to the final forms in which it is used by enzymes in both the producing organisms and other species, including humans who acquire it through their diet.
Radical SAM is a designation for a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. rSAMs comprise the largest superfamily of metal-containing enzymes.
2-deoxy-scyllo-inosamine dehydrogenase (SAM-dependent) is an enzyme with systematic name 2-deoxy-scyllo-inosamine:S-adenosyl-L-methionine 1-oxidoreductase. This enzyme catalyses the following chemical reaction
23S rRNA (adenosine1067-2'-O)-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (adenosine1067-2'-O)-methyltransferase. This enzyme catalyses the following chemical reaction
DTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose N,N-dimethyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:dTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose 3-N,N-dimethyltransferase. This enzyme catalyses the following chemical reaction
DTDP-3-amino-3,6-dideoxy-alpha-D-galactopyranose N,N-dimethyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:dTDP-3-amino-3,6-dideoxy-alpha-D-galactopyranose 3-N,N-dimethyltransferase. This enzyme catalyses the following chemical reaction
Trans-resveratrol di-O-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:trans-resveratrol 3,5-O-dimethyltransferase. This enzyme catalyses the following chemical reaction
Geranyl diphosphate 2-C-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:geranyl-diphosphate 2-C-methyltransferase. This enzyme catalyses the following chemical reaction
DTDP-4-amino-4,6-dideoxy-D-galactose acyltransferase is an enzyme with systematic name acetyl-CoA:dTDP-4-amino-4,6-dideoxy-alpha-D-galactose N-acetyltransferase. This enzyme catalyses the following chemical reaction
Glycosyltransferase DesVII is an enzyme with systematic name dTDP-3-dimethylamino-3,4,6-trideoxy-alpha-D-glucopyranose:10-deoxymethynolide 3-dimethylamino-4,6-dideoxy-alpha-D-glucosyltransferase. This enzyme catalyses the following chemical reaction
Desosaminyl transferase EryCIII is an enzyme with systematic name dTDP-3-dimethylamino-4,6-dideoxy-alpha-D-glucopyranose:3-alpha-mycarosylerythronolide B 3-dimethylamino-4,6-dideoxy-alpha-D-glucosyltransferase. This enzyme catalyses the following chemical reaction
S-Adenosylmethionine:tRNA ribosyltransferase-isomerase is an enzyme with systematic name S-adenosyl-L-methionine:7-aminomethyl-7-deazaguanosine ribosyltransferase . This enzyme catalyses the following chemical reaction
DTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose transaminase is an enzyme with systematic name dTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose:2-oxoglutarate aminotransferase. This enzyme catalyses the following chemical reaction
DTDP-3-amino-3,6-dideoxy-alpha-D-galactopyranose transaminase is an enzyme with systematic name dTDP-3-amino-3,6-dideoxy-alpha-D-galactopyranose:2-oxoglutarate aminotransferase. This enzyme catalyses the following chemical reaction
Phosphomethylpyrimidine synthase is an enzyme with systematic name 5-amino-1-(5-phospho-D-ribosyl)imidazole formate-lyase . This enzyme catalyses the following chemical reaction
TDP-4-oxo-6-deoxy-alpha-D-glucose-3,4-oxoisomerase (dTDP-3-dehydro-6-deoxy-alpha-D-glucopyranose-forming) is an enzyme with systematic name dTDP-4-dehydro-6-deoxy-alpha-D-glucopyranose:dTDP-3-dehydro-6-deoxy-alpha-D-glucopyranose isomerase. This enzyme catalyses the following chemical reaction






