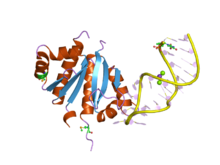
Nucleoproteins are proteins conjugated with nucleic acids. Typical nucleoproteins include ribosomes, nucleosomes and viral nucleocapsid proteins.

Heterogeneous nuclear ribonucleoproteins (hnRNPs) are complexes of RNA and protein present in the cell nucleus during gene transcription and subsequent post-transcriptional modification of the newly synthesized RNA (pre-mRNA). The presence of the proteins bound to a pre-mRNA molecule serves as a signal that the pre-mRNA is not yet fully processed and therefore not ready for export to the cytoplasm. Since most mature RNA is exported from the nucleus relatively quickly, most RNA-binding protein in the nucleus exist as heterogeneous ribonucleoprotein particles. After splicing has occurred, the proteins remain bound to spliced introns and target them for degradation.

In molecular biology, LSm proteins are a family of RNA-binding proteins found in virtually every cellular organism. LSm is a contraction of 'like Sm', because the first identified members of the LSm protein family were the Sm proteins. LSm proteins are defined by a characteristic three-dimensional structure and their assembly into rings of six or seven individual LSm protein molecules, and play a large number of various roles in mRNA processing and regulation.

rRNA 2'-O-methyltransferase fibrillarin is an enzyme that in humans is encoded by the FBL gene.

Heterogeneous nuclear ribonucleoprotein A1 is a protein that in humans is encoded by the HNRNPA1 gene. Mutations in hnRNP A1 are causative of amyotrophic lateral sclerosis and the syndrome multisystem proteinopathy.

Heterogeneous nuclear ribonucleoprotein K is a protein that in humans is encoded by the HNRNPK gene. It is found in the cell nucleus that binds to pre-messenger RNA (mRNA) as a component of heterogeneous ribonucleoprotein particles. The simian homolog is known as protein H16. Both proteins bind to single-stranded DNA as well as to RNA and can stimulate the activity of RNA polymerase II, the protein responsible for most gene transcription. The relative affinities of the proteins for DNA and RNA vary with solution conditions and are inversely correlated, so that conditions promoting strong DNA binding result in weak RNA binding.

Heterogeneous nuclear ribonucleoproteins A2/B1 is a protein that in humans is encoded by the HNRNPA2B1 gene.

Heterogeneous nuclear ribonucleoprotein U is a protein that in humans is encoded by the HNRNPU gene.

Poly(rC)-binding protein 1 is a protein that in humans is encoded by the PCBP1 gene.

Poly(rC)-binding protein 2 is a protein that in humans is encoded by the PCBP2 gene.
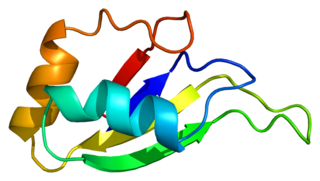
Heterogeneous nuclear ribonucleoprotein D0 (HNRNPD) also known as AU-rich element RNA-binding protein 1 (AUF1) is a protein that in humans is encoded by the HNRNPD gene. Alternative splicing of this gene results in four transcript variants.

Heterogeneous nuclear ribonucleoproteins C1/C2 is a protein that in humans is encoded by the HNRNPC gene.
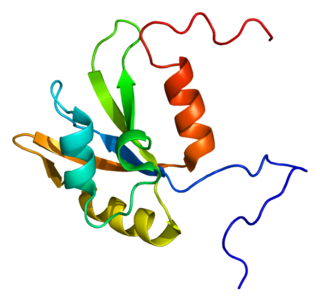
Heterogeneous nuclear ribonucleoprotein F is a protein that in humans is encoded by the HNRNPF gene.
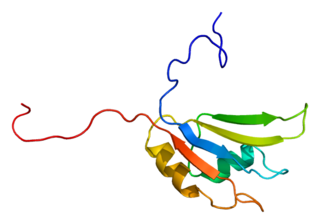
Heterogeneous nuclear ribonucleoprotein H is a protein that in humans is encoded by the HNRNPH1 gene.

Heterogeneous nuclear ribonucleoprotein A/B, also known as HNRNPAB, is a protein which in humans is encoded by the HNRNPAB gene. Although this gene is named HNRNPAB in reference to its first cloning as an RNA binding protein with similarity to HNRNP A and HNRNP B, it is not a member of the HNRNP A/B subfamily of HNRNPs, but groups together closely with HNRNPD/AUF1 and HNRNPDL.

Polypyrimidine tract-binding protein 1 is a protein that in humans is encoded by the PTBP1 gene.

Prp24 is a protein part of the pre-messenger RNA splicing process and aids the binding of U6 snRNA to U4 snRNA during the formation of spliceosomes. Found in eukaryotes from yeast to E. coli, fungi, and humans, Prp24 was initially discovered to be an important element of RNA splicing in 1989. Mutations in Prp24 were later discovered in 1991 to suppress mutations in U4 that resulted in cold-sensitive strains of yeast, indicating its involvement in the reformation of the U4/U6 duplex after the catalytic steps of splicing.

RNA recognition motif, RNP-1 is a putative RNA-binding domain of about 90 amino acids that are known to bind single-stranded RNAs. It was found in many eukaryotic proteins.
The RNA-binding Proteins Database (RBPDB) is a biological database of RNA-binding protein specificities that includes experimental observations of RNA-binding sites. The experimental results included are both in vitro and in vivo from primary literature. It includes four metazoan species, which are Homo sapiens, Mus musculus, Drosophila melanogaster, and Caenorhabditis elegans. RNA-binding domains included in this database are RNA recognition motif, K homology, CCCH zinc finger, and more domains. As of 2021, the latest RBPDB release includes 1,171 RNA-binding proteins.
The arginine-glycine or arginine-glycine-glycine (RG/RGG) motif is a repeating amino acid sequence motif commonly found in RNA-binding proteins (RBPs). RGG regions in proteins are defined as two or more RG/RGG sequences within a stretch of 30 amino acids. Initially named the RGG box, it confers a protein with the ability to bind double-stranded mRNA molecules. The RGG motif has been observed in proteins from at least 12 animal species, including humans.