
Evolutionary developmental biology is a field of biological research that compares the developmental processes of different organisms to infer the ancestral relationships between them and how developmental processes evolved.

Molecular evolution is the process of change in the sequence composition of cellular molecules such as DNA, RNA, and proteins across generations. The field of molecular evolution uses principles of evolutionary biology and population genetics to explain patterns in these changes. Major topics in molecular evolution concern the rates and impacts of single nucleotide changes, neutral evolution vs. natural selection, origins of new genes, the genetic nature of complex traits, the genetic basis of speciation, evolution of development, and ways that evolutionary forces influence genomic and phenotypic changes.
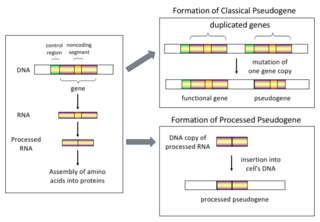
Pseudogenes, sometimes referred to as zombie genes in the media, are segments of DNA that are related to real genes. Pseudogenes have lost at least some functionality, relative to the complete gene, in cellular gene expression or protein-coding ability. Pseudogenes often result from the accumulation of multiple mutations within a gene whose product is not required for the survival of the organism, but can also be caused by genomic copy number variation (CNV) where segments of 1+ kb are duplicated or deleted. Although not fully functional, pseudogenes may be functional, similar to other kinds of noncoding DNA, which can perform regulatory functions. The "pseudo" in "pseudogene" implies a variation in sequence relative to the parent coding gene, but does not necessarily indicate pseudo-function. Despite being non-coding, many pseudogenes have important roles in normal physiology and abnormal pathology.

A generegulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins. These play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo).

Evolutionary biology is the subfield of biology that studies the evolutionary processes that produced the diversity of life on Earth, starting from a single common ancestor. These processes include natural selection, common descent, and speciation.

A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Different polypeptide chains may have different functions. This is distinct from a multienzyme complex, in which multiple catalytic domains are found in a single polypeptide chain.

This is a list of topics in evolutionary biology.

The theory of facilitated variation demonstrates how seemingly complex biological systems can arise through a limited number of regulatory genetic changes, through the differential re-use of pre-existing developmental components. The theory was presented in 2005 by Marc W. Kirschner and John C. Gerhart.
Computational neurogenetic modeling (CNGM) is concerned with the study and development of dynamic neuronal models for modeling brain functions with respect to genes and dynamic interactions between genes. These include neural network models and their integration with gene network models. This area brings together knowledge from various scientific disciplines, such as computer and information science, neuroscience and cognitive science, genetics and molecular biology, as well as engineering.

A biological network is any network that applies to biological systems. A network is any system with sub-units that are linked into a whole, such as species units linked into a whole food web. Biological networks provide a mathematical representation of connections found in ecological, evolutionary, and physiological studies, such as neural networks. The analysis of biological networks with respect to human diseases has led to the field of network medicine.
Morphogenetic robotics generally refers to the methodologies that address challenges in robotics inspired by biological morphogenesis.

Günter P. Wagner is Alison Richard Professor of Ecology and Evolutionary biology at Yale University, and head of the Wagner Lab.
Natural computing, also called natural computation, is a terminology introduced to encompass three classes of methods: 1) those that take inspiration from nature for the development of novel problem-solving techniques; 2) those that are based on the use of computers to synthesize natural phenomena; and 3) those that employ natural materials to compute. The main fields of research that compose these three branches are artificial neural networks, evolutionary algorithms, swarm intelligence, artificial immune systems, fractal geometry, artificial life, DNA computing, and quantum computing, among others.

Wagner's gene network model is a computational model of artificial gene networks, which explicitly modeled the developmental and evolutionary process of genetic regulatory networks. A population with multiple organisms can be created and evolved from generation to generation. It was first developed by Andreas Wagner in 1996 and has been investigated by other groups to study the evolution of gene networks, gene expression, robustness, plasticity and epistasis.
Evolution by gene duplication is an event by which a gene or part of a gene can have two identical copy that can not be distinguished from each other. This phenomenon is understood to be an important source of novelty in evolution, providing for an expanded repertoire of molecular activities. The underlying mutational event of duplication may be a conventional gene duplication mutation within a chromosome, or a larger-scale event involving whole chromosomes (aneuploidy) or whole genomes (polyploidy). A classic view, owing to Susumu Ohno, which is known as Ohno model, he explains how duplication creates redundancy, the redundant copy accumulates beneficial mutations which provides fuel for innovation. Knowledge of evolution by gene duplication has advanced more rapidly in the past 15 years due to new genomic data, more powerful computational methods of comparative inference, and new evolutionary models.

Robustness of a biological system is the persistence of a certain characteristic or trait in a system under perturbations or conditions of uncertainty. Robustness in development is known as canalization. According to the kind of perturbation involved, robustness can be classified as mutational, environmental, recombinational, or behavioral robustness etc. Robustness is achieved through the combination of many genetic and molecular mechanisms and can evolve by either direct or indirect selection. Several model systems have been developed to experimentally study robustness and its evolutionary consequences.
Albert Erives is a developmental geneticist who studies transcriptional enhancers underlying animal development and diseases of development (cancers). Erives also proposed the pacRNA model for the dual origin of the genetic code and universal homochirality. He is known for work at the intersection of genetics, evolution, developmental biology, and gene regulation. He has worked at the California Institute of Technology, University of California, Berkeley, and Dartmouth College, and is an associate professor at the University of Iowa.

Andreas Wagner is an Austrian/US evolutionary biologist and professor at the University of Zürich, Switzerland. He is known for his work on the role of robustness and innovation in biological evolution. Wagner is professor and chairman at the Department of Evolutionary Biology and Environmental Studies at the University of Zürich.

The following outline is provided as an overview of and topical guide to evolution:
In evolutionary biology, developmental bias refers to the production against or towards certain ontogenetic trajectories which ultimately influence the direction and outcome of evolutionary change by affecting the rates, magnitudes, directions and limits of trait evolution. Historically, the term was synonymized with developmental constraint, however, the latter has been more recently interpreted as referring solely to the negative role of development in evolution.

















