
A homeobox is a DNA sequence, around 180 base pairs long, that regulates large-scale anatomical features in the early stages of embryonic development. Mutations in a homeobox may change large-scale anatomical features of the full-grown organism.
Segmentation in biology is the division of some animal and plant body plans into a linear series of repetitive segments that may or may not be interconnected to each other. This article focuses on the segmentation of animal body plans, specifically using the examples of the taxa Arthropoda, Chordata, and Annelida. These three groups form segments by using a "growth zone" to direct and define the segments. While all three have a generally segmented body plan and use a growth zone, they use different mechanisms for generating this patterning. Even within these groups, different organisms have different mechanisms for segmenting the body. Segmentation of the body plan is important for allowing free movement and development of certain body parts. It also allows for regeneration in specific individuals.

The tusk shells or tooth shells, technically the Scaphopoda, are members of a class of shelled marine mollusc with worldwide distribution, and are the only class of exclusively infaunal marine molluscs. Shells of species within this class range in length 0.5–18 cm (0.20–7.09 in). Members of the order Dentaliida tend to be larger than those of the order Gadilida.
The ParaHox gene cluster is an array of homeobox genes from the Gsx, Xlox (Pdx) and Cdx gene families.
Hox genes, a subset of homeobox genes, are a group of related genes that specify regions of the body plan of an embryo along the head-tail axis of animals. Hox proteins encode and specify the characteristics of 'position', ensuring that the correct structures form in the correct places of the body. For example, Hox genes in insects specify which appendages form on a segment, and Hox genes in vertebrates specify the types and shape of vertebrae that will form. In segmented animals, Hox proteins thus confer segmental or positional identity, but do not form the actual segments themselves.

T-box transcription factor T, also known as Brachyury protein, is encoded for in humans by the TBXT gene. Brachyury functions as a transcription factor within the T-box family of genes. Brachyury homologs have been found in all bilaterian animals that have been screened, as well as the freshwater cnidarian Hydra.
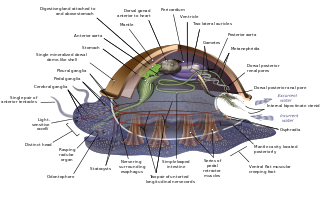
The evolution of the molluscs is the way in which the Mollusca, one of the largest groups of invertebrate animals, evolved. This phylum includes gastropods, bivalves, scaphopods, cephalopods, and several other groups. The fossil record of mollusks is relatively complete, and they are well represented in most fossil-bearing marine strata. Very early organisms which have dubiously been compared to molluscs include Kimberella and Odontogriphus.

The mir-10 microRNA precursor is a short non-coding RNA gene involved in gene regulation. It is part of an RNA gene family which contains mir-10, mir-51, mir-57, mir-99 and mir-100. mir-10, mir-99 and mir-100 have now been predicted or experimentally confirmed in a wide range of species. miR-51 and miR-57 have currently only been identified in the nematode Caenorhabditis elegans.
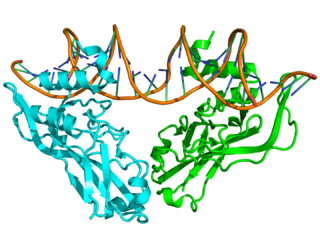
T-box refers to a group of transcription factors involved in embryonic limb and heart development. Every T-box protein has a relatively large DNA-binding domain, generally comprising about a third of the entire protein that is both necessary and sufficient for sequence-specific DNA binding. All members of the T-box gene family bind to the "T-box", a DNA consensus sequence of TCACACCT.

The (pan)arthropod head problem is a long-standing zoological dispute concerning the segmental composition of the heads of the various arthropod groups, and how they are evolutionarily related to each other. While the dispute has historically centered on the exact make-up of the insect head, it has been widened to include other living arthropods, such as chelicerates, myriapods, and crustaceans, as well as fossil forms, such as the many arthropods known from exceptionally preserved Cambrian faunas. While the topic has classically been based on insect embryology, in recent years a great deal of developmental molecular data has become available. Dozens of more or less distinct solutions to the problem, dating back to at least 1897, have been published, including several in the 2000s.

PDX1, also known as insulin promoter factor 1, is a transcription factor in the ParaHox gene cluster. In vertebrates, Pdx1 is necessary for pancreatic development, including β-cell maturation, and duodenal differentiation. In humans this protein is encoded by the PDX1 gene, which was formerly known as IPF1. The gene was originally identified in the clawed frog Xenopus laevis and is present widely across the evolutionary diversity of bilaterian animals, although it has been lost in evolution in arthropods and nematodes. Despite the gene name being Pdx1, there is no Pdx2 gene in most animals; single-copy Pdx1 orthologs have been identified in all mammals. Coelacanth and cartilaginous fish are, so far, the only vertebrates shown to have two Pdx genes, Pdx1 and Pdx2.

Homeobox protein Hox-A3 is a protein that in humans is encoded by the HOXA3 gene.

Marine invertebrates are the invertebrates that live in marine habitats. Invertebrate is a blanket term that includes all animals apart from the vertebrate members of the chordate phylum. Invertebrates lack a vertebral column, and some have evolved a shell or a hard exoskeleton. As on land and in the air, marine invertebrates have a large variety of body plans, and have been categorised into over 30 phyla. They make up most of the macroscopic life in the oceans.

The molluscshell is typically a calcareous exoskeleton which encloses, supports and protects the soft parts of an animal in the phylum Mollusca, which includes snails, clams, tusk shells, and several other classes. Not all shelled molluscs live in the sea; many live on the land and in freshwater.

Mollusca is the second-largest phylum of invertebrate animals, after Arthropoda; members are known as molluscs or mollusks. Around 76,000 extant species of molluscs are recognized. The number of fossil species is estimated between 60,000 and 100,000 additional species. The proportion of undescribed species is very high. Many taxa remain poorly studied.
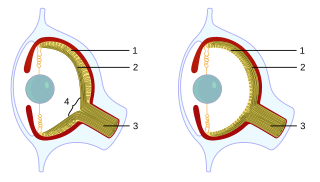
Cephalopods, as active marine predators, possess sensory organs specialized for use in aquatic conditions. They have a camera-type eye which consists of an iris, a circular lens, vitreous cavity, pigment cells, and photoreceptor cells that translate light from the light-sensitive retina into nerve signals which travel along the optic nerve to the brain. For the past 140 years, the camera-type cephalopod eye has been compared with the vertebrate eye as an example of convergent evolution, where both types of organisms have independently evolved the camera-eye trait and both share similar functionality. Contention exists on whether this is truly convergent evolution or parallel evolution. Unlike the vertebrate camera eye, the cephalopods' form as invaginations of the body surface, and consequently the cornea lies over the top of the eye as opposed to being a structural part of the eye. Unlike the vertebrate eye, a cephalopod eye is focused through movement, much like the lens of a camera or telescope, rather than changing shape as the lens in the human eye does. The eye is approximately spherical, as is the lens, which is fully internal.

Euperipatoides kanangrensis is a species of velvet worm of the Peripatopsidae family, described in 1996 from specimens collected in Kanangra-Boyd National Park, New South Wales. This species has 15 pairs of legs in both sexes. It is endemic to Australia. The embryonic development of Euperipatoideskanangrensis has been described. This species is viviparous. This species is used as model organism for the last common ancestor of the Panarthropoda. It resembles fossil Cambrian lobopodians.

Euperipatoides rowelli is an ovoviviparous species of velvet worm of the Peripatopsidae family. It is found in New South Wales and the Australian Capital Territory.
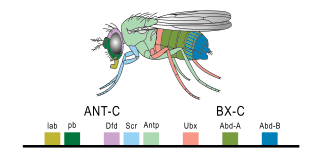
The evo-devo gene toolkit is the small subset of genes in an organism's genome whose products control the organism's embryonic development. Toolkit genes are central to the synthesis of molecular genetics, palaeontology, evolution and developmental biology in the science of evolutionary developmental biology (evo-devo). Many of them are ancient and highly conserved among animal phyla.
The protocerebrum is the first segment of the panarthropod brain.
















