Drosophila is a genus of fly, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many species to linger around overripe or rotting fruit. They should not be confused with the Tephritidae, a related family, which are also called fruit flies ; tephritids feed primarily on unripe or ripe fruit, with many species being regarded as destructive agricultural pests, especially the Mediterranean fruit fly.
Microevolution is the change in allele frequencies that occurs over time within a population. This change is due to four different processes: mutation, selection, gene flow and genetic drift. This change happens over a relatively short amount of time compared to the changes termed macroevolution.
Speciation is the evolutionary process by which populations evolve to become distinct species. The biologist Orator F. Cook coined the term in 1906 for cladogenesis, the splitting of lineages, as opposed to anagenesis, phyletic evolution within lineages. Charles Darwin was the first to describe the role of natural selection in speciation in his 1859 book On the Origin of Species. He also identified sexual selection as a likely mechanism, but found it problematic.

Theodosius Grigorievich Dobzhansky was an American geneticist and evolutionary biologist. He was a central figure in the field of evolutionary biology for his work in shaping the modern synthesis and also popular for his support and promotion of theistic evolution as a practicing Christian. Born in the Russian Empire, Dobzhansky immigrated to the United States in 1927, aged 27.
Allopatric speciation – also referred to as geographic speciation, vicariant speciation, or its earlier name the dumbbell model – is a mode of speciation that occurs when biological populations become geographically isolated from each other to an extent that prevents or interferes with gene flow.

Haldane's rule is an observation about the early stage of speciation, formulated in 1922 by the British evolutionary biologist J. B. S. Haldane, that states that if — in a species hybrid — only one sex is inviable or sterile, that sex is more likely to be the heterogametic sex. The heterogametic sex is the one with two different sex chromosomes; in therian mammals, for example, this is the male.
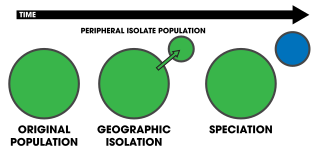
Peripatric speciation is a mode of speciation in which a new species is formed from an isolated peripheral population. Since peripatric speciation resembles allopatric speciation, in that populations are isolated and prevented from exchanging genes, it can often be difficult to distinguish between them., and peripatric speciation may be considered one type or model of allopatric speciation. The primary distinguishing characteristic of peripatric speciation is that one of the populations is much smaller than the other, as opposed to allopatric speciation, in which similarly-sized populations become separated. The terms peripatric and peripatry are often used in biogeography, referring to organisms whose ranges are closely adjacent but do not overlap, being separated where these organisms do not occur—for example on an oceanic island compared to the mainland. Such organisms are usually closely related ; their distribution being the result of peripatric speciation.

Genetics and the Origin of Species is a 1937 book by the Ukrainian-American evolutionary biologist Theodosius Dobzhansky. It is regarded as one of the most important works of modern synthesis and was one of the earliest. The book popularized the work of population genetics to other biologists and influenced their appreciation for the genetic basis of evolution. In his book, Dobzhansky applied the theoretical work of Sewall Wright (1889–1988) to the study of natural populations, allowing him to address evolutionary problems in a novel way during his time. Dobzhansky implements theories of mutation, natural selection, and speciation throughout his book to explain the habits of populations and the resulting effects on their genetic behavior. The book explains evolution in depth as a process over time that accounts for the diversity of all life on Earth. The study of evolution was present, but greatly neglected at the time. Dobzhansky illustrates that evolution regarding the origin and nature of species during this time in history was deemed mysterious, but had expanding potential for progress to be made in its field.

In evolutionary biology, disruptive selection, also called diversifying selection, describes changes in population genetics in which extreme values for a trait are favored over intermediate values. In this case, the variance of the trait increases and the population is divided into two distinct groups. In this more individuals acquire peripheral character value at both ends of the distribution curve.

In parapatric speciation, two subpopulations of a species evolve reproductive isolation from one another while continuing to exchange genes. This mode of speciation has three distinguishing characteristics: 1) mating occurs non-randomly, 2) gene flow occurs unequally, and 3) populations exist in either continuous or discontinuous geographic ranges. This distribution pattern may be the result of unequal dispersal, incomplete geographical barriers, or divergent expressions of behavior, among other things. Parapatric speciation predicts that hybrid zones will often exist at the junction between the two populations.
The mechanisms of reproductive isolation are a collection of evolutionary mechanisms, behaviors and physiological processes critical for speciation. They prevent members of different species from producing offspring, or ensure that any offspring are sterile. These barriers maintain the integrity of a species by reducing gene flow between related species.

Drosophila pseudoobscura is a species of fruit fly, used extensively in lab studies of speciation. It is native to western North America.

Ecological speciation is a form of speciation arising from reproductive isolation that occurs due to an ecological factor that reduces or eliminates gene flow between two populations of a species. Ecological factors can include changes in the environmental conditions in which a species experiences, such as behavioral changes involving predation, predator avoidance, pollinator attraction, and foraging; as well as changes in mate choice due to sexual selection or communication systems. Ecologically-driven reproductive isolation under divergent natural selection leads to the formation of new species. This has been documented in many cases in nature and has been a major focus of research on speciation for the past few decades.
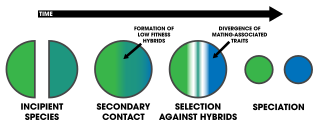
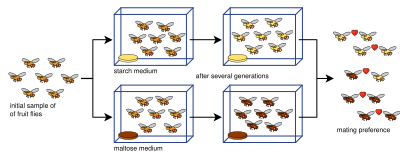
Reinforcement is a process of speciation where natural selection increases the reproductive isolation between two populations of species. This occurs as a result of selection acting against the production of hybrid individuals of low fitness. The idea was originally developed by Alfred Russel Wallace and is sometimes referred to as the Wallace effect. The modern concept of reinforcement originates from Theodosius Dobzhansky. He envisioned a species separated allopatrically, where during secondary contact the two populations mate, producing hybrids with lower fitness. Natural selection results from the hybrid's inability to produce viable offspring; thus members of one species who do not mate with members of the other have greater reproductive success. This favors the evolution of greater prezygotic isolation. Reinforcement is one of the few cases in which selection can favor an increase in prezygotic isolation, influencing the process of speciation directly. This aspect has been particularly appealing among evolutionary biologists.

The scientific study of speciation — how species evolve to become new species — began around the time of Charles Darwin in the middle of the 19th century. Many naturalists at the time recognized the relationship between biogeography and the evolution of species. The 20th century saw the growth of the field of speciation, with major contributors such as Ernst Mayr researching and documenting species' geographic patterns and relationships. The field grew in prominence with the modern evolutionary synthesis in the early part of that century. Since then, research on speciation has expanded immensely.

Reinforcement is a process within speciation where natural selection increases the reproductive isolation between two populations of species by reducing the production of hybrids. Evidence for speciation by reinforcement has been gathered since the 1990s, and along with data from comparative studies and laboratory experiments, has overcome many of the objections to the theory. Differences in behavior or biology that inhibit formation of hybrid zygotes are termed prezygotic isolation. Reinforcement can be shown to be occurring by measuring the strength of prezygotic isolation in a sympatric population in comparison to an allopatric population of the same species. Comparative studies of this allow for determining large-scale patterns in nature across various taxa. Mating patterns in hybrid zones can also be used to detect reinforcement. Reproductive character displacement is seen as a result of reinforcement, so many of the cases in nature express this pattern in sympatry. Reinforcement's prevalence is unknown, but the patterns of reproductive character displacement are found across numerous taxa, and is considered to be a common occurrence in nature. Studies of reinforcement in nature often prove difficult, as alternative explanations for the detected patterns can be asserted. Nevertheless, empirical evidence exists for reinforcement occurring across various taxa and its role in precipitating speciation is conclusive.

The Drosophila quinaria species group is a speciose lineage of mushroom-feeding flies studied for their specialist ecology, their parasites, population genetics, and the evolution of immune systems. Quinaria species are part of the Drosophila subgenus.
Maria R. Servedio is a Canadian-American professor at the University of North Carolina at Chapel Hill. Her research spans a wide range of topics in evolutionary biology from sexual selection to evolution of behavior. She largely approaches these topics using mathematical models. Her current research interests include speciation and reinforcement, mate choice, and learning with a particular focus on evolutionary mechanisms that promote premating (prezygotic) isolation. Through integrative approaches and collaborations, she uses mathematical models along with experimental, genetic, and comparative techniques to draw conclusions on how evolution occurs. She has published extensively on these topics and has more than 50 peer-reviewed articles. She served as Vice President in 2018 of the American Society of Naturalists, and has been elected to serve as President in 2023.
Allochronic speciation is a form of speciation arising from reproductive isolation that occurs due to a change in breeding time that reduces or eliminates gene flow between two populations of a species. The term allochrony is used to describe the general ecological phenomenon of the differences in phenology that arise between two or more species—speciation caused by allochrony is effectively allochronic speciation.
In biology, parallel speciation is a type of speciation where there is repeated evolution of reproductively isolating traits via the same mechanisms occurring between separate yet closely related species inhabiting different environments. This leads to a circumstance where independently evolved lineages have developed reproductive isolation from their ancestral lineage, but not from other independent lineages that inhabit similar environments. In order for parallel speciation to be confirmed, there is a set of three requirements that has been established that must be met: there must be phylogenetic independence between the separate populations inhabiting similar environments to ensure that the traits responsible for reproductive isolation evolved separately, there must be reproductive isolation not only between the ancestral population and the descendent population, but also between descendent populations that inhabit dissimilar environments, and descendent populations that inhabit similar environments must not be reproductively isolated from one another. To determine if natural selection specifically is the cause of parallel speciation, a fourth requirement has been established that includes identifying and testing an adaptive mechanism, which eliminates the possibility of a genetic factor such as polyploidy being the responsible agent.