A restriction enzyme, restriction endonuclease, or restrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.

In biochemistry, the DNA methyltransferase family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor.
The restriction modification system is found in bacteria and other prokaryotic organisms, and provides a defense against foreign DNA, such as that borne by bacteriophages.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltrasferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the nucleophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases.
Site-specific DNA-methyltransferase (cytosine-N4-specific) is an enzyme with systematic name S-adenosyl-L-methionine:DNA-cytosine N4-methyltransferase. This enzyme catalyses the following chemical reaction
Type I site-specific deoxyribonuclease is an enzyme. This enzyme catalyses the following chemical reaction
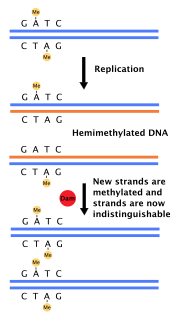
DAM methylase, an abbreviation for deoxyadenosine methylase, is an enzyme that adds a methyl group to the adenine of the sequence 5'-GATC-3' in newly synthesized DNA. Immediately after DNA synthesis, the daughter strand remains unmethylated for a short time. It is an orphan methyltransferase that is not part of a restriction-modification system and regulates gene expression. Dam methylase is unique to prokaryotes and bacteriophages.
DamID is a molecular biology protocol used to map the binding sites of DNA- and chromatin-binding proteins in eukaryotes. DamID identifies binding sites by expressing the proposed DNA-binding protein as a fusion protein with DNA methyltransferase. Binding of the protein of interest to DNA localizes the methyltransferase in the region of the binding site. Adenosine methylation does not occur naturally in eukaryotes and therefore adenine methylation in any region can be concluded to have been caused by the fusion protein, implying the region is located near a binding site. DamID is an alternate method to ChIP-on-chip or ChIP-seq.
PstI is a type II restriction endonuclease isolated from the Gram negative species, Providencia stuartii.

DNA base flipping, or nucleotide flipping, is a mechanism in which a single nucleotide base, or nucleobase, is rotated outside the nucleic acid double helix. This occurs when a nucleic acid-processing enzyme needs access to the base to perform work on it, such as its excision for replacement with another base during DNA repair. It was first observed in 1994 using X-ray crystallography in a methyltransferase enzyme catalyzing methylation of a cytosine base in DNA. Since then, it has been shown to be used by different enzymes in many biological processes such as DNA methylation, various DNA repair mechanisms, and DNA replication. It can also occur in RNA double helices or in the DNA:RNA intermediates formed during RNA transcription.
CcrM is an orphan DNA Methyltransferase, that is involved in controlling gene expression in most Alphaproteobacteria. This enzyme modifies DNA by catalyzing the transference of a methyl group from the S-adenosyl-L methionine substrate to the N6 position of an adenine base in the sequence 5'-GANTC-3' with high specificity. In Caulobacter crescentus Ccrm is produced at the end of the replication cycle when Ccrm recognition sites are hemimethylated, rapidly methylating the DNA. CcrM is essential in other Alphaproteobacteria but is role is not yet determined. CcrM is a highly specific methyltransferase with a novel DNA recognition mechanism.




