
Transfer RNA is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length, that serves as the physical link between the mRNA and the amino acid sequence of proteins. Transfer RNA (tRNA) does this by carrying an amino acid to the protein synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code.
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.

Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease.

RNA editing is a molecular process through which some cells can make discrete changes to specific nucleotide sequences within an RNA molecule after it has been generated by RNA polymerase. It occurs in all living organisms and is one of the most evolutionarily conserved properties of RNAs. RNA editing may include the insertion, deletion, and base substitution of nucleotides within the RNA molecule. RNA editing is relatively rare, with common forms of RNA processing not usually considered as editing. It can affect the activity, localization as well as stability of RNAs, and has been linked with human diseases.

Dihydrolipoyl transacetylase is an enzyme component of the multienzyme pyruvate dehydrogenase complex. The pyruvate dehydrogenase complex is responsible for the pyruvate decarboxylation step that links glycolysis to the citric acid cycle. This involves the transformation of pyruvate from glycolysis into acetyl-CoA which is then used in the citric acid cycle to carry out cellular respiration.
Aspartate—tRNAAsn ligase is an enzyme with systematic name L-aspartate:tRNAAsx ligase (AMP-forming). This enzyme catalyses the following chemical reaction
In enzymology, a threonine-tRNA ligase is an enzyme that catalyzes the chemical reaction
Tyrosine—tRNA ligase, also known as tyrosyl-tRNA synthetase is an enzyme that is encoded by the gene YARS. Tyrosine—tRNA ligase catalyzes the chemical reaction

Histone acetyltransferase KAT2A is an enzyme that in humans is encoded by the KAT2A gene.
CCA tRNA nucleotidyltransferase is an enzyme with systematic name CTP,CTP,ATP:tRNA cytidylyl,cytidylyl,adenylyltransferase. This enzyme catalyses the following chemical reaction
Histone acetyltransferase KAT5 is an enzyme that in humans is encoded by the KAT5 gene. It is also commonly identified as TIP60.

Elongator complex protein 3, also named KAT9, is a protein that in humans is encoded by the ELP3 gene. ELP3 is the catalytic histone acetyltransferase subunit of the RNA polymerase II elongator complex, which is a component of the RNA polymerase II holoenzyme and is involved in transcriptional elongation. ELP3 supports the migration and branching of projection neurons through acetylation of alpha-tubulin in the developing cerebral cortex. In mammals, ELP3 is important for paternal DNA demethylation after fertilization. ELP3 is potentially involved in cellular redox homeostasis by mediating the acetylation of glucose-6-phosphate dehydrogenase. Besides, ELP3 may play a role in chromatin remodeling and is involved in acetylation of histones H3 and probably H4.

K(lysine) acetyltransferase 8 (KAT8) is an enzyme that in humans is encoded by the KAT8 gene.

Heparan-α-glucosaminide N-acetyltransferase is an enzyme that in humans is encoded by the HGSNAT gene.
Lysidine is an uncommon nucleoside, rarely seen outside of tRNA. It is a derivative of cytidine in which the carbonyl is replaced by the amino acid lysine. The third position in the anti-codon of the Isoleucine-specific tRNA, is typically changed from a cytidine which would pair with guanosine to a lysidine which will base pair with adenosine. Uridine could not be used at this position even though it is a conventional partner for adenosine since it will also "wobble base pair" with guanosine. So lysidine allows better translation fidelity. Lysidine is denoted as L or k2C.

Acetyl-CoA acetyltransferase, cytosolic, also known as cytosolic acetoacetyl-CoA thiolase, is an enzyme that in humans is encoded by the ACAT2 gene

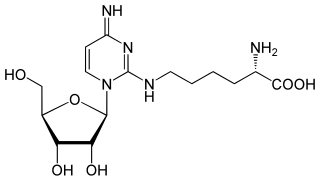
Agmatidine (2-agmatinylcytidine, symbol C+ or agm2C) is a modified cytidine present in the wobble position of the anticodon of several archaeal AUA decoding tRNAs. Agmatidine is essential for correct decoding of the AUA codon in many archaea and is required for aminoacylation of tRNAIle2 with isoleucine.

In biochemistry, wybutosine (yW) is a heavily modified nucleoside of phenylalanine transfer RNA that stabilizes interactions between the codons and anti-codons during protein synthesis. Ensuring accurate synthesis of protein is essential in maintaining health as defects in tRNA modifications are able to cause disease. In eukaryotic organisms, it is found only in position 37, 3'-adjacent to the anticodon, of phenylalanine tRNA. Wybutosine enables correct translation through the stabilization of the codon-anticodon base pairing during the decoding process.
tRNA (guanine37-N1)-methyltransferase (EC 2.1.1.228, TrmD, tRNA (m1G37) methyltransferase, transfer RNA (m1G37) methyltransferase, Trm5p, TRMT5, tRNA-(N1G37) methyltransferase, MJ0883 (gene)) is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (guanine37-N1)-methyltransferase. This enzyme catalyses the following chemical reaction
TRNAIle-lysidine synthase (EC 6.3.4.19, TilS, mesJ (gene), yacA (gene), isoleucine-specific transfer ribonucleate lysidine synthetase, tRNAIle-lysidine synthetase) is an enzyme with systematic name L-lysine:(tRNAIle2)-cytidine34 ligase (AMP-forming). This enzyme catalyses the following chemical reaction












