
Pseudomonas putida is a Gram-negative, rod-shaped, saprophytic soil bacterium. It has a versatile metabolism and is amenable to genetic manipulation, making it a common organism used in research, bioremediation, and synthesis of chemicals and other compounds.
4-Hydroxybenzoic acid, also known as p-hydroxybenzoic acid (PHBA), is a monohydroxybenzoic acid, a phenolic derivative of benzoic acid. It is a white crystalline solid that is slightly soluble in water and chloroform but more soluble in polar organic solvents such as alcohols and acetone. 4-Hydroxybenzoic acid is primarily known as the basis for the preparation of its esters, known as parabens, which are used as preservatives in cosmetics and some ophthalmic solutions. It is isomeric with 2-hydroxybenzoic acid, known as salicylic acid, a precursor to aspirin, and with 3-hydroxybenzoic acid.

Muconate lactonizing enzymes are involved in the breakdown of lignin-derived aromatics, catechol and protocatechuate, to citric acid cycle intermediates as a part of the β-ketoadipate pathway in soil microbes. Some bacterial species are also capable of dehalogenating chloroaromatic compounds by the action of chloromuconate lactonizing enzymes. MLEs consist of several strands which have variable reaction favorable parts therefore the configuration of the strands affect its ability to accept protons. The bacterial MLEs belong to the enolase superfamily, several structures from which are known. MLEs have an identifying structure made up of two proteins and two Magnesium ions as well as various classes depending on whether it is bacterial or eukaryotic. The reaction mechanism that MLEs undergo are the reverse of beta-elimination in which the enolate alpha-carbon is protonated. MLEs can undergo mutations caused by a deletion of catB structural genes which can cause some bacteria to lose its functions such as the ability to grow. Additional mutations to MLEs can cause its structure and function to alter and could cause the conformation to change therefore making it an inactive enzyme that is unable to bind its substrate. There is another enzyme called Mandelate Racemase that is very similar to MLEs in the structural way as well as them both being a part of the enolase superfamily. They both have the same end product even though they undergo different chemical reactions in order to reach the end product.
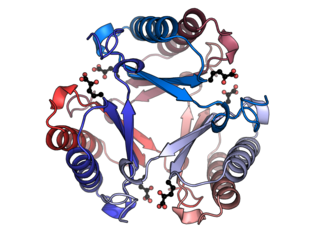
4-Oxalocrotonate tautomerase or 4-OT is an enzyme that converts 2-hydroxymuconate to the αβ-unsaturated ketone, 2-oxo-3-hexenedioate. This enzyme forms part of a bacterial metabolic pathway that oxidatively catabolizes toluene, o-xylene, 3-ethyltoluene, and 1,2,4-trimethylbenzene into intermediates of the citric acid cycle. With a monomer size of just 62 amino acid residues, the 4-Oxalocrotonate tautomerase is one of the smallest enzyme subunits known. However, in solution, the enzyme forms a hexamer of six identical subunits, so the active site may be formed by amino acid residues from several subunits. This enzyme is also unusual in that it uses a proline residue at the amino terminus as an active site residue.

In enzymology, a homoisocitrate dehydrogenase (EC 1.1.1.87) is an enzyme that catalyzes the chemical reaction
In enzymology, a quinoline 2-oxidoreductase (EC 1.3.99.17) is an enzyme that catalyzes the chemical reaction
In enzymology, a 4-methoxybenzoate monooxygenase (O-demethylating) (EC 1.14.99.15) is an enzyme that catalyzes the chemical reaction
In enzymology, a 3-carboxy-cis,cis-muconate cycloisomerase is an enzyme that catalyzes the chemical reaction
In enzymology, a muconolactone Δ-isomerase is an enzyme that catalyzes the chemical reaction
The enzyme 4-oxalmesaconate hydratase (EC 4.2.1.83) catalyzes the chemical reaction
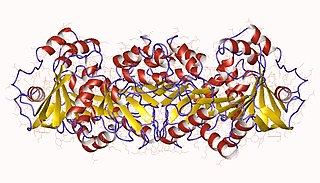
The enzyme homoaconitate hydratase (EC 4.2.1.36) catalyzes the chemical reaction
The enzyme 2-pyrone-4,6-dicarboxylate lactonase (EC 3.1.1.57, LigI) catalyzes the reversible hydrolytic reaction
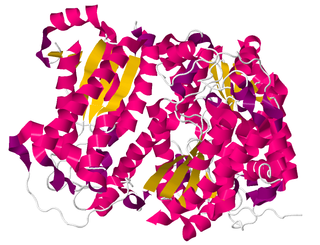
In enzymology, a creatininase (EC 3.5.2.10) is an enzyme that catalyses the hydrolysis of creatinine to creatine, which can then be metabolised to urea and sarcosine by creatinase.

In enzymology, a homocitrate synthase (EC 2.3.3.14) is an enzyme that catalyzes the chemical reaction
Gallate dioxygenase (EC 1.13.11.57, GalA) is an enzyme with systematic name gallate:oxygen oxidoreductase. This enzyme catalyses the following chemical reaction
2,5-diketocamphane 1,2-monooxygenase (EC 1.14.14.108, 2,5-diketocamphane lactonizing enzyme, ketolactonase I, 2,5-diketocamphane 1,2-monooxygenase oxygenating component, 2,5-DKCMO, camphor 1,2-monooxygenase, camphor ketolactonase I) is an enzyme with systematic name (+)-bornane-2,5-dione,NADH:oxygen oxidoreductase (1,2-lactonizing). This enzyme catalyses the following chemical reaction

Nicotinate dehydrogenase (cytochrome) (EC 1.17.2.1, nicotinic acid hydroxylase, nicotinate hydroxylase) is an enzyme with systematic name nicotinate:cytochrome 6-oxidoreductase (hydroxylating). This enzyme catalyses the following chemical reaction
Lupanine 17-hydroxylase (cytochrome c) (EC 1.17.2.2, lupanine dehydrogenase (cytochrome c)) is an enzyme with systematic name lupanine:cytochrome c-oxidoreductase (17-hydroxylating). This enzyme catalyses the following chemical reaction
Putidaredoxin—NAD+ reductase (EC 1.18.1.5, putidaredoxin reductase, camA (gene)) is an enzyme with systematic name putidaredoxin:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
Methanogen homoaconitase (EC 4.2.1.114, methanogen HACN) is an enzyme with systematic name (R)-2-hydroxybutane-1,2,4-tricarboxylate hydro-lyase ((1R,2S)-1-hydroxybutane-1,2,4-tricarboxylate-forming). This enzyme catalyses the following chemical reaction








