| prostaglandin-I synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC no. | 5.3.99.4 | ||||||||
| CAS no. | 65802-86-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
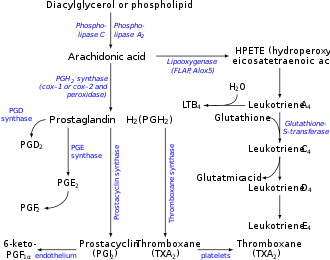
Prostaglandin-I synthase (EC 5.3.99.4) also known as prostaglandin I2 (prostacyclin) synthase (PTGIS) or CYP8A1 is an enzyme involved in prostanoid biosynthesis that in humans is encoded by the PTGIS gene. [4] This enzyme belongs to the family of cytochrome P450 isomerases.