Related Research Articles

Transfer RNA is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length. In a cell, it provides the physical link between the genetic code in messenger RNA (mRNA) and the amino acid sequence of proteins, carrying the correct sequence of amino acids to be combined by the protein-synthesizing machinery, the ribosome. Each three-nucleotide codon in mRNA is complemented by a three-nucleotide anticodon in tRNA. As such, tRNAs are a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code.
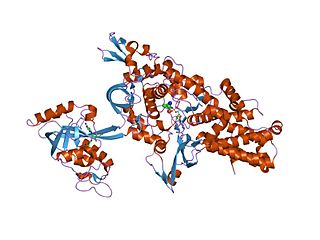
An aminoacyl-tRNA synthetase, also called tRNA-ligase, is an enzyme that attaches the appropriate amino acid onto its corresponding tRNA. It does so by catalyzing the transesterification of a specific cognate amino acid or its precursor to one of all its compatible cognate tRNAs to form an aminoacyl-tRNA. In humans, the 20 different types of aa-tRNA are made by the 20 different aminoacyl-tRNA synthetases, one for each amino acid of the genetic code.

EF-Tu is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochondria as TUFM.

In enzymology, an arginine—tRNA ligase is an enzyme that catalyzes the chemical reaction
Aspartate—tRNAAsn ligase is an enzyme with systematic name L-aspartate:tRNAAsx ligase (AMP-forming). This enzyme catalyses the following chemical reaction
In enzymology, a glutamate—tRNAGln ligase is an enzyme that catalyzes the chemical reaction
In enzymology, a glutamate—tRNA ligase is an enzyme that catalyzes the chemical reaction
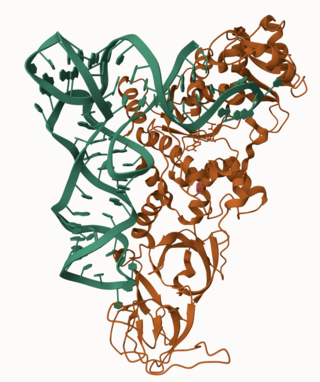
Glutamine—tRNA ligase or glutaminyl-tRNA synthetase (GlnRS) is an aminoacyl-tRNA synthetase, also called tRNA-ligase. is an enzyme that attaches the amino acid glutamine onto its cognate tRNA.

In enzymology, a phenylalanine—tRNA ligase is an enzyme that catalyzes the chemical reaction
In enzymology, a threonine-tRNA ligase is an enzyme that catalyzes the chemical reaction
Tyrosine—tRNA ligase, also known as tyrosyl-tRNA synthetase is an enzyme that is encoded by the gene YARS. Tyrosine—tRNA ligase catalyzes the chemical reaction
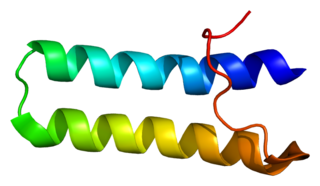
Bifunctional aminoacyl-tRNA synthetase is an enzyme that in humans is encoded by the EPRS gene.

Glutaminyl-tRNA synthetase is an enzyme that in humans is encoded by the QARS gene.
Amino acid activation refers to the attachment of an amino acid to its respective transfer RNA (tRNA). The reaction occurs in the cell cytosol and consists of two steps: first, the enzyme aminoacyl tRNA synthetase catalyzes the binding of adenosine triphosphate (ATP) to a corresponding amino acid, forming a reactive aminoacyl adenylate intermediate and releasing inorganic pyrophosphate (PPi). Subsequently, aminoacyl tRNA synthetase binds the AMP-amino acid to a tRNA molecule, releasing AMP and attaching the amino acid to the tRNA. The resulting aminoacyl-tRNA is said to be charged.
Aminoacyl-tRNA synthetases, class II is a family of proteins. These proteins catalyse the attachment of an amino acid to its cognate transfer RNA molecule in a highly specific two-step reaction. These proteins differ widely in size and oligomeric state, and have a limited sequence homology.

EF-P is an essential protein that in bacteria stimulates the formation of the first peptide bonds in protein synthesis. Studies show that EF-P prevents ribosomes from stalling during the synthesis of proteins containing consecutive prolines. EF-P binds to a site located between the binding site for the peptidyl tRNA and the exiting tRNA. It spans both ribosomal subunits with its amino-terminal domain positioned adjacent to the aminoacyl acceptor stem and its carboxyl-terminal domain positioned next to the anticodon stem-loop of the P site-bound initiator tRNA. The EF-P protein shape and size is very similar to a tRNA and interacts with the ribosome via the exit “E” site on the 30S subunit and the peptidyl-transferase center (PTC) of the 50S subunit. EF-P is a translation aspect of an unknown function, therefore It probably functions indirectly by altering the affinity of the ribosome for aminoacyl-tRNA, thus increasing their reactivity as acceptors for peptidyl transferase.

The B3/B4 domain, is found in tRNA synthetase beta subunits, as well as in some non-tRNA synthetase proteins.

In molecular biology, Yce-I protein domain is a putative periplasmic protein. This entry represents the lipid-binding protein YceI from Escherichia coli and the polyisoprenoid-binding protein TTHA0802 from Thermus thermophilus. Its role is to help aid the biosynthesis of isoprenoid, an important molecule found in all living organisms.

In molecular biology, YqeY is a type of protein domain of unknown function. It is thought to have a role in protein synthesis, facilitating the production of charged transfer RNA used in the process of translating mRNA into protein. It is present as a domain of glutaminyl-tRNA synthetase (GlnRS) in almost all eukaryotes.
Dino Moras, born on 23 November 1944, is a French biochemist, research director at the CNRS and co-director of the Institute of Genetics and Molecular and Cellular Biology (IGBMC) in Illkirch-Graffenstaden until 2010.
References
- ↑ Delarue M, Moras D, Poch O, Eriani G, Gangloff J (1990). "Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs". Nature. 347 (6289): 203–206. Bibcode:1990Natur.347..203E. doi:10.1038/347203a0. PMID 2203971. S2CID 4324290.
- ↑ Moras D, Konno M, Shimada A, Nureki O, Tateno M, Yokoyama S, Sugiura I, Ugaji-Yoshikawa Y, Kuwabara S, Lorber B, Giege R (2000). "The 2.0 A crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules". Structure. 8 (2): 197–208. doi: 10.1016/S0969-2126(00)00095-2 . PMID 10673435.
- ↑ Perona JJ, Steitz TA, Rould MA (1993). "Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase". Biochemistry. 32 (34): 8758–8771. doi:10.1021/bi00085a006. PMID 8364025.
- ↑ Delarue M, Moras D (1993). "The aminoacyl-tRNA synthetase family: modules at work". BioEssays. 15 (10): 675–687. doi:10.1002/bies.950151007. PMID 8274143. S2CID 35612984.
- ↑ Schimmel P (1991). "Classes of aminoacyl-tRNA synthetases and the establishment of the genetic code". Trends Biochem. Sci. 16 (1): 1–3. doi:10.1016/0968-0004(91)90002-D. PMID 2053131.
- ↑ Cusack S, Leberman R, Hartlein M (1991). "Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases". Nucleic Acids Res. 19 (13): 3489–3498. doi:10.1093/nar/19.13.3489. PMC 328370 . PMID 1852601.
- ↑ Bairoch A (2004). "List of aminoacyl-tRNA synthetases".
{{cite journal}}: Cite journal requires|journal=(help) - ↑ Soll D, Freist W, Gauss DH, Lapointe J (1997). "Glutamyl-tRNA sythetase". Biol. Chem. 378 (11): 1313–1329. doi:10.1515/bchm.1997.378.11.1299. PMID 9426192.