
Systems biology is the computational and mathematical analysis and modeling of complex biological systems. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological systems, using a holistic approach to biological research.

Marc Wallace Kirschner is an American cell biologist and biochemist and the founding chair of the Department of Systems Biology at Harvard Medical School. He is known for major discoveries in cell and developmental biology related to the dynamics and function of the cytoskeleton, the regulation of the cell cycle, and the process of signaling in embryos, as well as the evolution of the vertebrate body plan. He is a leader in applying mathematical approaches to biology. He is the John Franklin Enders University Professor at Harvard University. In 2021 he was elected to the American Philosophical Society.
The Rat Genome Database (RGD) is a database of rat genomics, genetics, physiology and functional data, as well as data for comparative genomics between rat, human and mouse. RGD is responsible for attaching biological information to the rat genome via structured vocabulary, or ontology, annotations assigned to genes and quantitative trait loci (QTL), and for consolidating rat strain data and making it available to the research community. They are also developing a suite of tools for mining and analyzing genomic, physiologic and functional data for the rat, and comparative data for rat, mouse, human, and five other species.
Biomedical text mining refers to the methods and study of how text mining may be applied to texts and literature of the biomedical domain. As a field of research, biomedical text mining incorporates ideas from natural language processing, bioinformatics, medical informatics and computational linguistics. The strategies in this field have been applied to the biomedical literature available through services such as PubMed.

Tang Chao is a Chair Professor of Physics and Systems Biology at Peking University.

The Biological General Repository for Interaction Datasets (BioGRID) is a curated biological database of protein-protein interactions, genetic interactions, chemical interactions, and post-translational modifications created in 2003 (originally referred to as simply the General Repository for Interaction Datasets by Mike Tyers, Bobby-Joe Breitkreutz, and Chris Stark at the Lunenfeld-Tanenbaum Research Institute at Mount Sinai Hospital. It strives to provide a comprehensive curated resource for all major model organism species while attempting to remove redundancy to create a single mapping of data. Users of The BioGRID can search for their protein, chemical or publication of interest and retrieve annotation, as well as curated data as reported, by the primary literature and compiled by in house large-scale curation efforts. The BioGRID is hosted in Toronto, Ontario, Canada and Dallas, Texas, United States and is partnered with the Saccharomyces Genome Database, FlyBase, WormBase, PomBase, and the Alliance of Genome Resources. The BioGRID is funded by the NIH and CIHR. BioGRID is an observer member of the International Molecular Exchange Consortium.

BioModels is a free and open-source repository for storing, exchanging and retrieving quantitative models of biological interest created in 2006. All the models in the curated section of BioModels Database have been described in peer-reviewed scientific literature.

The Systems Biology Ontology (SBO) is a set of controlled, relational vocabularies of terms commonly used in systems biology, and in particular in computational modeling.
Xiaowei Zhuang is a Chinese-American biophysicist who is the David B. Arnold Jr. Professor of Science, Professor of Chemistry and Chemical Biology, and Professor of Physics at Harvard University, and an Investigator at the Howard Hughes Medical Institute. She is best known for her work in the development of Stochastic Optical Reconstruction Microscopy (STORM), a super-resolution fluorescence microscopy method, and the discoveries of novel cellular structures using STORM. She received a 2019 Breakthrough Prize in Life Sciences for developing super-resolution imaging techniques that get past the diffraction limits of traditional light microscopes, allowing scientists to visualize small structures within living cells. She was elected a Member of the American Philosophical Society in 2019 and was awarded a Vilcek Foundation Prize in Biomedical Science in 2020.

Uri Alon is a Professor and Systems Biologist at the Weizmann Institute of Science. His highly cited research investigates gene expression, network motifs and the design principles of biological networks in Escherichia coli and other organisms using both computational biology and traditional experimental wet laboratory techniques.

SABIO-RK is a web-accessible database storing information about biochemical reactions and their kinetic properties.
Heng Li is a Chinese bioinformatics scientist. He is an associate professor at the department of Biomedical Informatics of Harvard Medical School and the department of Data Science of Dana-Farber Cancer Institute. He was previously a research scientist working at the Broad Institute in Cambridge, Massachusetts with David Reich and David Altshuler. Li's work has made several important contributions in the field of next generation sequencing.
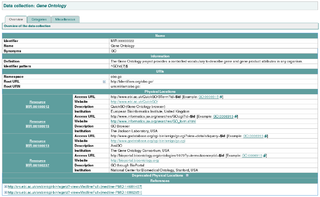
The MIRIAM Registry, a by-product of the MIRIAM Guidelines, is a database of namespaces and associated information that is used in the creation of uniform resource identifiers. It contains the set of community-approved namespaces for databases and resources serving, primarily, the biological sciences domain. These shared namespaces, when combined with 'data collection' identifiers, can be used to create globally unique identifiers for knowledge held in data repositories. For more information on the use of URIs to annotate models, see the specification of SBML Level 2 Version 2.

Ron Milo is a Professor of Systems Biology at the Weizmann Institute of Science. He is Weizmann Dean of Education, the chairperson of the Israel society of ecology and environmental sciences and the director of the Institute for environmental sustainability at Weizmann. Formerly he was the chairperson of the Israel young academy.

Galit Lahav is an Israeli-American systems biologist and Professor of Systems Biology at Harvard Medical School. In 2018 she became Chair of the Department of Systems Biology at Harvard Medical School. She is known for discovering the pulsatile behavior of the tumor suppressor protein p53 and uncovering its significance for cell fate, and for her contributions to the culture of mentoring in science. She lives in Boston, Massachusetts.
Biocuration is the field of life sciences dedicated to organizing biomedical data, information and knowledge into structured formats, such as spreadsheets, tables and knowledge graphs. The biocuration of biomedical knowledge is made possible by the cooperative work of biocurators, software developers and bioinformaticians and is at the base of the work of biological databases.

Arren Bar-Even (6 June 1980 – 18 September 2020) was an Israeli biochemist and synthetic biologist. In his research, Bar-Even made pioneering advances in the design and implementation of novel pathways for improved CO2 fixation. and formate utilisation.
Jasmin Fisher, is an Israeli-British biologist who is Professor of computational biology at University College London. She is Group Leader of the Fisher Lab at UCL Cancer Institute, which develops state-of-the-art computational models and analysis techniques to study cancer evolution and mechanisms of drug resistance to identify better personalised treatments for cancer patients.
Jennifer Waters is an American scientist who is a Lecturer on Cell Biology, the Director of the Core for Imaging Technology & Education(CITE; formally the NIC) and the Director of the Cell Biology Microscopy Facility at Harvard Medical School. She is an imaging expert and educator whose efforts to educate life scientists about microscopy and to systemize the education of microscopists in microscopy facilities serve as a blueprint for similar efforts worldwide.
Matt Thomson is an American computational biologist, academic, and entrepreneur. He works in the fields of computational biology, biophysics, and machine learning.











