Related Research Articles

Microsoft Access is a database management system (DBMS) from Microsoft that combines the relational Access Database Engine (ACE) with a graphical user interface and software-development tools. It is a member of the Microsoft 365 suite of applications, included in the Professional and higher editions or sold separately.
Waveform Audio File Format is an audio file format standard for storing an audio bitstream on personal computers. The format was developed and published for the first time in 1991 by IBM and Microsoft. It is the main format used on Microsoft Windows systems for uncompressed audio. The usual bitstream encoding is the linear pulse-code modulation (LPCM) format.
Resource Interchange File Format (RIFF) is a generic file container format for storing data in tagged chunks. It is primarily used for audio and video, though it can be used for arbitrary data.
In bioinformatics, sequence analysis is the process of subjecting a DNA, RNA or peptide sequence to any of a wide range of analytical methods to understand its features, function, structure, or evolution. It can be performed on the entire genome, transcriptome or proteome of an organism, and can also involve only selected segments or regions, like tandem repeats and transposable elements. Methodologies used include sequence alignment, searches against biological databases, and others.

General transcription factors (GTFs), also known as basal transcriptional factors, are a class of protein transcription factors that bind to specific sites (promoter) on DNA to activate transcription of genetic information from DNA to messenger RNA. GTFs, RNA polymerase, and the mediator constitute the basic transcriptional apparatus that first bind to the promoter, then start transcription. GTFs are also intimately involved in the process of gene regulation, and most are required for life.
Microsoft Assistance Markup Language is an XML-based markup language developed by the Microsoft User Assistance Platform team to provide user assistance for the Microsoft Windows Vista operating system. It makes up the Assistance Platform on Windows Vista.
GTF may stand for:
In bioinformatics, the general feature format is a file format used for describing genes and other features of DNA, RNA and protein sequences.
Transcription factor TFIIA is a nuclear protein involved in the RNA polymerase II-dependent transcription of DNA. TFIIA is one of several general (basal) transcription factors (GTFs) that are required for all transcription events that use RNA polymerase II. Other GTFs include TFIID, a complex composed of the TATA binding protein TBP and TBP-associated factors (TAFs), as well as the factors TFIIB, TFIIE, TFIIF, and TFIIH. Together, these factors are responsible for promoter recognition and the formation of a transcription preinitiation complex (PIC) capable of initiating RNA synthesis from a DNA template.
Hunk is the executable file format of tools and programs of the Amiga Operating System based on Motorola 68000 CPU and other processors of the same family. The file format was originally defined by MetaComCo. as part of TRIPOS, which formed the basis for AmigaDOS. This kind of executable got its name from the fact that the software programmed on Amiga is divided in its internal structure into many pieces called hunks, in which every portion could contain either code or data.
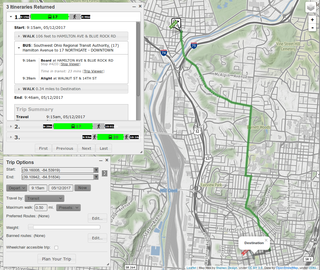
A journey planner, trip planner, or route planner is a specialized search engine used to find an optimal means of travelling between two or more given locations, sometimes using more than one transport mode. Searches may be optimized on different criteria, for example fastest, shortest, fewest changes, cheapest. They may be constrained, for example, to leave or arrive at a certain time, to avoid certain waypoints, etc. A single journey may use a sequence of several modes of transport, meaning the system may know about public transport services as well as transport networks for private transportation. Trip planning or journey planning is sometimes distinguished from route planning, which is typically thought of as using private modes of transportation such as cycling, driving, or walking, normally using a single mode at a time. Trip or journey planning, in contrast, would make use of at least one public transport mode which operates according to published schedules; given that public transport services only depart at specific times, an algorithm must therefore not only find a path to a destination, but seek to optimize it so as to minimize the waiting time incurred for each leg. In European Standards such as Transmodel, trip planning is used specifically to describe the planning of a route for a passenger, to avoid confusion with the completely separate process of planning the operational journeys to be made by public transport vehicles on which such trips are made.
A file format is a standard way that information is encoded for storage in a computer file. It specifies how bits are used to encode information in a digital storage medium. File formats may be either proprietary or free.

UGENE is computer software for bioinformatics. It works on personal computer operating systems such as Windows, macOS, or Linux. It is released as free and open-source software, under a GNU General Public License (GPL) version 2.
GENCODE is a scientific project in genome research and part of the ENCODE scale-up project.

Integrated Genome Browser (IGB) is an open-source genome browser, a visualization tool used to observe biologically-interesting patterns in genomic data sets, including sequence data, gene models, alignments, and data from DNA microarrays.
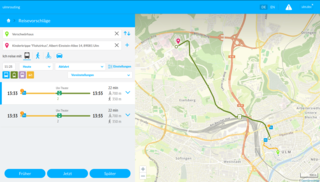
GTFS, which stands for General Transit Feed Specification or (originally) Google Transit Feed Specification, defines a common format for public transportation schedules and associated geographic information. GTFS contains only static or scheduled information about public transport services, and is sometimes known as GTFS Static or GTFS Schedule to distinguish it from the GTFS Realtime extension, which defines how information on the realtime status of services can be shared.
Ensembl Genomes is a scientific project to provide genome-scale data from non-vertebrate species.
GFF may refer to:
The BED format is a text file format used to store genomic regions as coordinates and associated annotations. The data are presented in the form of columns separated by spaces or tabs. This format was developed during the Human Genome Project and then adopted by other sequencing projects. As a result of this increasingly wide use, this format had already become a de facto standard in bioinformatics before a formal specification was written.
References
- ↑ GFF/GTF info, from Ensembl