
Coenzyme A (CoA, SHCoA, CoASH) is a coenzyme, notable for its role in the synthesis and oxidation of fatty acids, and the oxidation of pyruvate in the citric acid cycle. All genomes sequenced to date encode enzymes that use coenzyme A as a substrate, and around 4% of cellular enzymes use it (or a thioester) as a substrate. In humans, CoA biosynthesis requires cysteine, pantothenate (vitamin B5), and adenosine triphosphate (ATP).

Acetyl-CoA carboxylase (ACC) is a biotin-dependent enzyme that catalyzes the irreversible carboxylation of acetyl-CoA to produce malonyl-CoA through its two catalytic activities, biotin carboxylase (BC) and carboxyltransferase (CT). ACC is a multi-subunit enzyme in most prokaryotes and in the chloroplasts of most plants and algae, whereas it is a large, multi-domain enzyme in the cytoplasm of most eukaryotes. The most important function of ACC is to provide the malonyl-CoA substrate for the biosynthesis of fatty acids. The activity of ACC can be controlled at the transcriptional level as well as by small molecule modulators and covalent modification. The human genome contains the genes for two different ACCs—ACACA and ACACB.

Malonyl-CoA is a coenzyme A derivative of malonic acid.

Oxaloacetate decarboxylase is a carboxy-lyase involved in the conversion of oxaloacetate into pyruvate.

In molecular biology, Beta-ketoacyl-ACP synthase EC 2.3.1.41, is an enzyme involved in fatty acid synthesis. It typically uses malonyl-CoA as a carbon source to elongate ACP-bound acyl species, resulting in the formation of ACP-bound β-ketoacyl species such as acetoacetyl-ACP.
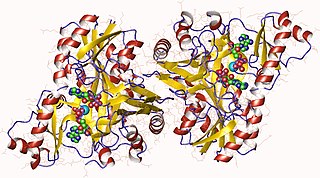
In enzymology, a biotin carboxylase (EC 6.3.4.14) is an enzyme that catalyzes the chemical reaction
In enzymology, a [acyl-carrier-protein] S-malonyltransferase is an enzyme that catalyzes the chemical reaction
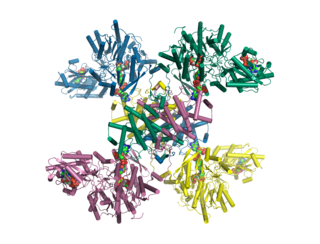
ATP citrate synthase (also ATP citrate lyase (ACLY)) is an enzyme that in animals represents an important step in fatty acid biosynthesis. By converting citrate to acetyl-CoA, the enzyme links carbohydrate metabolism, which yields citrate as an intermediate, with fatty acid biosynthesis, which consumes acetyl-CoA. In plants, ATP citrate lyase generates cytosolic acetyl-CoA precursors of thousands of specialized metabolites, including waxes, sterols, and polyketides.

Fatty-acyl-CoA Synthase, or more commonly known as yeast fatty acid synthase, is an enzyme complex responsible for fatty acid biosynthesis, and is of Type I Fatty Acid Synthesis (FAS). Yeast fatty acid synthase plays a pivotal role in fatty acid synthesis. It is a 2.6 MDa barrel shaped complex and is composed of two, unique multi-functional subunits: alpha and beta. Together, the alpha and beta units are arranged in an α6β6 structure. The catalytic activities of this enzyme complex involves a coordination system of enzymatic reactions between the alpha and beta subunits. The enzyme complex therefore consists of six functional centers for fatty acid synthesis.
Malonyl-S-ACP:biotin-protein carboxyltransferase is an enzyme with systematic name malonyl-(acyl-carrier protein):biotinyl-(protein) carboxytransferase. This enzyme catalyses the following chemical reaction
Acetyl-S-ACP:malonate ACP transferase is an enzyme with systematic name acetyl-(acyl-carrier-protein):malonate S-(acyl-carrier-protein)transferase. This enzyme catalyses the following chemical reaction
Malonate decarboxylase holo-(acyl-carrier protein) synthase is an enzyme with systematic name 2'-(5-triphosphoribosyl)-3'-dephospho-CoA:apo-malonate-decarboxylase 2'-(5-phosphoribosyl)-3'-dephospho-CoA-transferase . This enzyme catalyses the following chemical reaction
The enzyme Pimelyl-[acyl-carrier protein] methyl ester esterase (EC 3.1.1.85, BioH; systematic name pimelyl-[acyl-carrier protein] methyl ester hydrolase catalyses the reaction
Malonyl-S-ACP decarboxylase (EC 4.1.1.87, malonyl-S-acyl-carrier protein decarboxylase, MdcD/MdcE, MdcD,E) is an enzyme with systematic name malonyl-(acyl-carrier-protein) carboxy-lyase. This enzyme catalyses the following chemical reaction
Biotin-independent malonate decarboxylase (EC 4.1.1.88, malonate decarboxylase (without biotin), malonate decarboxylase, MDC) is an enzyme with systematic name malonate carboxy-lyase (biotin-independent). This enzyme catalyses the following chemical reaction
Carboxybiotin decarboxylase (EC 7.2.4.1, MadB, carboxybiotin protein decarboxylase) is an enzyme with systematic name carboxybiotinyl-(protein) carboxy-lyase. This enzyme catalyses the following chemical reaction
Acetate—[acyl-carrier protein] ligase is an enzyme with systematic name acetate:(acyl-carrier-protein) ligase (AMP-forming). This enzyme catalyses the following chemical reaction
The Malonate Uptake (MatC) family is a constituent of the ion transporter (IT) superfamily. It consists of proteins from Gram-negative and Gram-positive bacteria, simple eukaryotes and archaea. The proteins are of about 450 amino acyl residues in length with 12-14 putative transmembrane segments (TMSs). Closest functionally-characterized homologues are in the DASS family. One member of this family is a putative malonate transporter.
The Malonate:Na+ Symporter (MSS) Family (TC# 2.A.70) is a group of transport proteins belonging to the CPA superfamily. These proteins are composites with constituents ranging in size from 129 to 255 amino acyl residues (aas) and exhibiting 4 to 7 transmembrane segments (TMSs). A representative list of proteins belonging to the MSS family can be found in the Transporter Classification Database.
The Na+-transporting Carboxylic Acid Decarboxylase (NaT-DC) Family (TC# 3.B.1) is a family of porters that belong to the CPA superfamily. Members of this family have been characterized in both Gram-positive and Gram-negative bacteria. A representative list of proteins belonging to the NaT-DC family can be found in the Transporter Classification Database.








