In biology, taxonomy is the scientific study of naming, defining (circumscribing) and classifying groups of biological organisms based on shared characteristics. Organisms are grouped into taxa and these groups are given a taxonomic rank; groups of a given rank can be aggregated to form a more inclusive group of higher rank, thus creating a taxonomic hierarchy. The principal ranks in modern use are domain, kingdom, phylum, class, order, family, genus, and species. The Swedish botanist Carl Linnaeus is regarded as the founder of the current system of taxonomy, as he developed a ranked system known as Linnaean taxonomy for categorizing organisms and binomial nomenclature for naming organisms.

Daniel Hunt Janzen is an American evolutionary ecologist, and conservationist. He divides his time between his professorship in biology at the University of Pennsylvania, where he is the DiMaura Professor of Conservation Biology, and his research and field work in Costa Rica.

Telegonus fulgerator, the two-barred flasher, is a species of spread-wing skipper butterfly in the family Hesperiidae which may constitute a possible cryptic species complex. It ranges all over the Americas, from the southern United States to northern Argentina.
T. Ryan Gregory is a Canadian evolutionary biologist and genome biologist and a Professor of the Department of Integrative Biology and the Division of Genomic Diversity within the Biodiversity Institute of Ontario at the University of Guelph in Guelph, Ontario, Canada.

The Census of Marine Life was a 10-year, US $650 million scientific initiative, involving a global network of researchers in more than 80 nations, engaged to assess and explain the diversity, distribution, and abundance of life in the oceans. The world's first comprehensive Census of Marine Life — past, present, and future — was released in 2010 in London. Initially supported by funding from the Alfred P. Sloan Foundation, the project was successful in generating many times that initial investment in additional support and substantially increased the baselines of knowledge in often underexplored ocean realms, as well as engaging over 2,700 different researchers for the first time in a global collaborative community united in a common goal, and has been described as "one of the largest scientific collaborations ever conducted".


The NTNU University Museum in Trondheim is one of seven Norwegian university museums with natural and cultural history collections and exhibits. The museum has research and administrative responsibility over archaeology and biology in Central Norway. Additionally, the museum operates comprehensive community outreach programs and has exhibits in wooden buildings in Kalvskinnet.
A taxonomic database is a database created to hold information on biological taxa – for example groups of organisms organized by species name or other taxonomic identifier – for efficient data management and information retrieval. Taxonomic databases are routinely used for the automated construction of biological checklists such as floras and faunas, both for print publication and online; to underpin the operation of web-based species information systems; as a part of biological collection management ; as well as providing, in some cases, the taxon management component of broader science or biology information systems. They are also a fundamental contribution to the discipline of biodiversity informatics.

DNA barcoding is a method of species identification using a short section of DNA from a specific gene or genes. The premise of DNA barcoding is that by comparison with a reference library of such DNA sections, an individual sequence can be used to uniquely identify an organism to species, just as a supermarket scanner uses the familiar black stripes of the UPC barcode to identify an item in its stock against its reference database. These "barcodes" are sometimes used in an effort to identify unknown species or parts of an organism, simply to catalog as many taxa as possible, or to compare with traditional taxonomy in an effort to determine species boundaries.
The history of genetics can be represented on a timeline of events from the earliest work in the 1850s, to the DNA era starting in the 1940s, and the genomics era beginning in the 1970s.
The Barcode of Life Data System is a web platform specifically devoted to DNA barcoding. It is a cloud-based data storage and analysis platform developed at the Centre for Biodiversity Genomics in Canada. It consists of four main modules, a data portal, an educational portal, a registry of BINs, and a data collection and analysis workbench which provides an online platform for analyzing DNA sequences. Since its launch in 2005, BOLD has been extended to provide a range of functionality including data organization, validation, visualization and publication. The most recent version of the system, version 4, launched in 2017, brings a set of improvements supporting data collection and analysis but also includes novel functionality improving data dissemination, citation, and annotation. Before November 16, 2020, BOLD already contained barcode sequences for 318,105 formally described species covering animals, plants, fungi, protists.

Sunil Kumar Verma, was an Indian biologist and a principal scientist at the Centre for Cellular and Molecular Biology, Hyderabad, India. Verma was primarily known for his contributions to the development of "universal primer technology", a first generation DNA barcoding method, that can identify any bird, fish, reptile or mammal from a small biological sample, and satisfy legal evidence requirements in a court of law. This technology has revitalised the field of wildlife forensics and is now routinely used across India to provide a species identification service in cases of wildlife crime. This approach of species identification is now known as "DNA barcoding" across the world.

Fungal Diversity Survey, or FunDiS, is a nonprofit citizen science organization formerly known as North American Mycoflora Project, Inc. FunDiS aims to document the diversity and distribution of fungi across North America “in order to increase awareness of their critical role in the health of ecosystems and allow us to better protect them in a world of rapid climate change and habitat loss.” The project encourages amateurs, working with professionals, to contribute observations to online databases vetted by experts, and to collect and document fungi for DNA barcoding. Fungal Diversity Survey, Inc. is a Charitable 501(c)(3) organization registered in Indiana, USA.

DNA barcoding is an alternative method to the traditional morphological taxonomic classification, and has frequently been used to identify species of aquatic macroinvertebrates. Many are crucial indicator organisms in the bioassessment of freshwater and marine ecosystems.

DNA barcoding of algae is commonly used for species identification and phylogenetic studies. Algae form a phylogenetically heterogeneous group, meaning that the application of a single universal barcode/marker for species delimitation is unfeasible, thus different markers/barcodes are applied for this aim in different algal groups.
Microbial DNA barcoding is the use of DNA metabarcoding to characterize a mixture of microorganisms. DNA metabarcoding is a method of DNA barcoding that uses universal genetic markers to identify DNA of a mixture of organisms.

DNA barcoding methods for fish are used to identify groups of fish based on DNA sequences within selected regions of a genome. These methods can be used to study fish, as genetic material, in the form of environmental DNA (eDNA) or cells, is freely diffused in the water. This allows researchers to identify which species are present in a body of water by collecting a water sample, extracting DNA from the sample and isolating DNA sequences that are specific for the species of interest. Barcoding methods can also be used for biomonitoring and food safety validation, animal diet assessment, assessment of food webs and species distribution, and for detection of invasive species.

Winifred Hallwachs is an American tropical ecologist who helped to establish and expand northwestern Costa Rica's Área de Conservación Guanacaste (ACG). The work of Hallwachs and her husband Daniel Janzen at ACG is considered an exemplar of inclusive conservation.

Fungal DNA barcoding is the process of identifying species of the biological kingdom Fungi through the amplification and sequencing of specific DNA sequences and their comparison with sequences deposited in a DNA barcode database such as the ISHAM reference database, or the Barcode of Life Data System (BOLD). In this attempt, DNA barcoding relies on universal genes that are ideally present in all fungi with the same degree of sequence variation. The interspecific variation, i.e., the variation between species, in the chosen DNA barcode gene should exceed the intraspecific (within-species) variation.
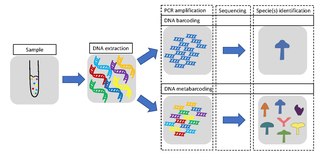
Metabarcoding is the barcoding of DNA/RNA in a manner that allows for the simultaneous identification of many taxa within the same sample. The main difference between barcoding and metabarcoding is that metabarcoding does not focus on one specific organism, but instead aims to determine species composition within a sample.









