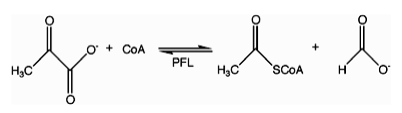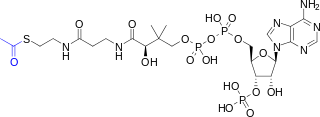
Acetyl-CoA is a molecule that participates in many biochemical reactions in protein, carbohydrate and lipid metabolism. Its main function is to deliver the acetyl group to the citric acid cycle to be oxidized for energy production. Coenzyme A consists of a β-mercaptoethylamine group linked to the vitamin pantothenic acid (B5) through an amide linkage and 3'-phosphorylated ADP. The acetyl group of acetyl-CoA is linked to the sulfhydryl substituent of the β-mercaptoethylamine group. This thioester linkage is a "high energy" bond, which is particularly reactive. Hydrolysis of the thioester bond is exergonic (−31.5 kJ/mol).
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.

In biochemistry, mixed acid fermentation is the metabolic process by which a six-carbon sugar is converted into a complex and variable mixture of acids. It is an anaerobic (non-oxygen-requiring) fermentation reaction that is common in bacteria. It is characteristic for members of the Enterobacteriaceae, a large family of Gram-negative bacteria that includes E. coli.
Oxidative decarboxylation is a decarboxylation reaction caused by oxidation. Most are accompanied by α- Ketoglutarate α- Decarboxylation caused by dehydrogenation of hydroxyl carboxylic acids such as carbonyl carboxylic acid, malic acid, isocitric acid, etc.
Pyruvate dehydrogenase is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. The conversion requires the coenzyme thiamine pyrophosphate.
Serine dehydratase or L-serine ammonia lyase (SDH) is in the β-family of pyridoxal phosphate-dependent (PLP) enzymes. SDH is found widely in nature, but its structural and properties vary among species. SDH is found in yeast, bacteria, and the cytoplasm of mammalian hepatocytes. SDH catalyzes is the deamination of L-serine to yield pyruvate, with the release of ammonia.
The enzyme cystathionine γ-lyase (EC 4.4.1.1, CTH or CSE; also cystathionase; systematic name L-cystathionine cysteine-lyase (deaminating; 2-oxobutanoate-forming)) breaks down cystathionine into cysteine, 2-oxobutanoate (α-ketobutyrate), and ammonia:
In enzymology, a [formate-C-acetyltransferase]-activating enzyme is an enzyme that catalyzes the chemical reaction
The enzyme 2-dehydro-3-deoxy-phosphogluconate aldolase, commonly known as KDPG aldolase, catalyzes the chemical reaction
The enzyme citramalyl-CoA lyase catalyzes the chemical reaction

In enzymology, an acetyl-CoA C-acetyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a [acyl-carrier-protein] S-acetyltransferase is an enzyme that catalyzes the reversible chemical reaction
In enzymology, a glycine C-acetyltransferase is an enzyme that catalyzes the chemical reaction:

In enzymology, a homocitrate synthase (EC 2.3.3.14) is an enzyme that catalyzes the chemical reaction

In enzymology, a serine O-acetyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a N-acetylneuraminate synthase (EC 2.5.1.56) is an enzyme that catalyzes the chemical reaction
Glyoxylate and dicarboxylate metabolism describes a variety of reactions involving glyoxylate or dicarboxylates. Glyoxylate is the conjugate base of glyoxylic acid, and within a buffered environment of known pH such as the cell cytoplasm these terms can be used almost interchangeably, as the gain or loss of a hydrogen ion is all that distinguishes them, and this can occur in the aqueous environment at any time. Likewise dicarboxylates are the conjugate bases of dicarboxylic acids, a general class of organic compounds containing two carboxylic acid groups, such as oxalic acid or succinic acid.
In molecular biology, the Cys/Met metabolism PLP-dependent enzyme family is a family of proteins including enzymes involved in cysteine and methionine metabolism which use PLP (pyridoxal-5'-phosphate) as a cofactor.
Radical SAM is a designation for a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. rSAMs comprise the largest superfamily of metal-containing enzymes.

Isethionate sulfite-lyase is a glycyl radical enzyme that catalyzes the degradation of isethionate into acetaldehyde and sulfite through the cleavage of a carbon-sulfur bond. This conversion is a necessary step for taurine catabolism in anaerobic bacteria like Bilophila wadsworthia. IslA is activated by the enzyme IslB which uses S-adenoslymethionine (SAM) as the initial radical donor.